SCHISM procedural example#
In this notebook we will use the SCHOSM grid, config and data objects to define a SCHISM workspace
Frontmatter#
Required inputs and defination of a few helper functions
[1]:
# Choose grid
# hgrid_file="hgrid.gr3" # heavy grid
hgrid_file="hgrid_20kmto60km_rompyschism_testing.gr3" # testing grid
[2]:
%load_ext autoreload
%autoreload 2
# turn off warnings
import warnings
warnings.filterwarnings('ignore')
from datetime import datetime
from pathlib import Path
from rompy.core import DataBlob, TimeRange
from shutil import rmtree
import xarray as xr
import numpy as np
import matplotlib.pyplot as plt
import intake
import cartopy.crs as ccrs
import pandas as pd
import logging
logging.basicConfig(level=logging.INFO)
HERE = Path('../../tests/schism')
Workspace basepath#
[3]:
workdir = Path("schism_procedural")
if workdir.exists():
rmtree(workdir)
workdir.mkdir(exist_ok=True)
Model Grid#
[4]:
# Grid object
from rompy.schism import Inputs, SCHISMGrid
#SCHISMGrid?
# Medium sized grid, will run one day in about 3 minutes on 48 cores
hgrid = HERE / "test_data" / hgrid_file
# Fast running grid, will run in about 1 minute on 4 cores
#hgrid = HERE / "test_data" / "hgrid_20kmto60km_rompyschism_testing.gr3"
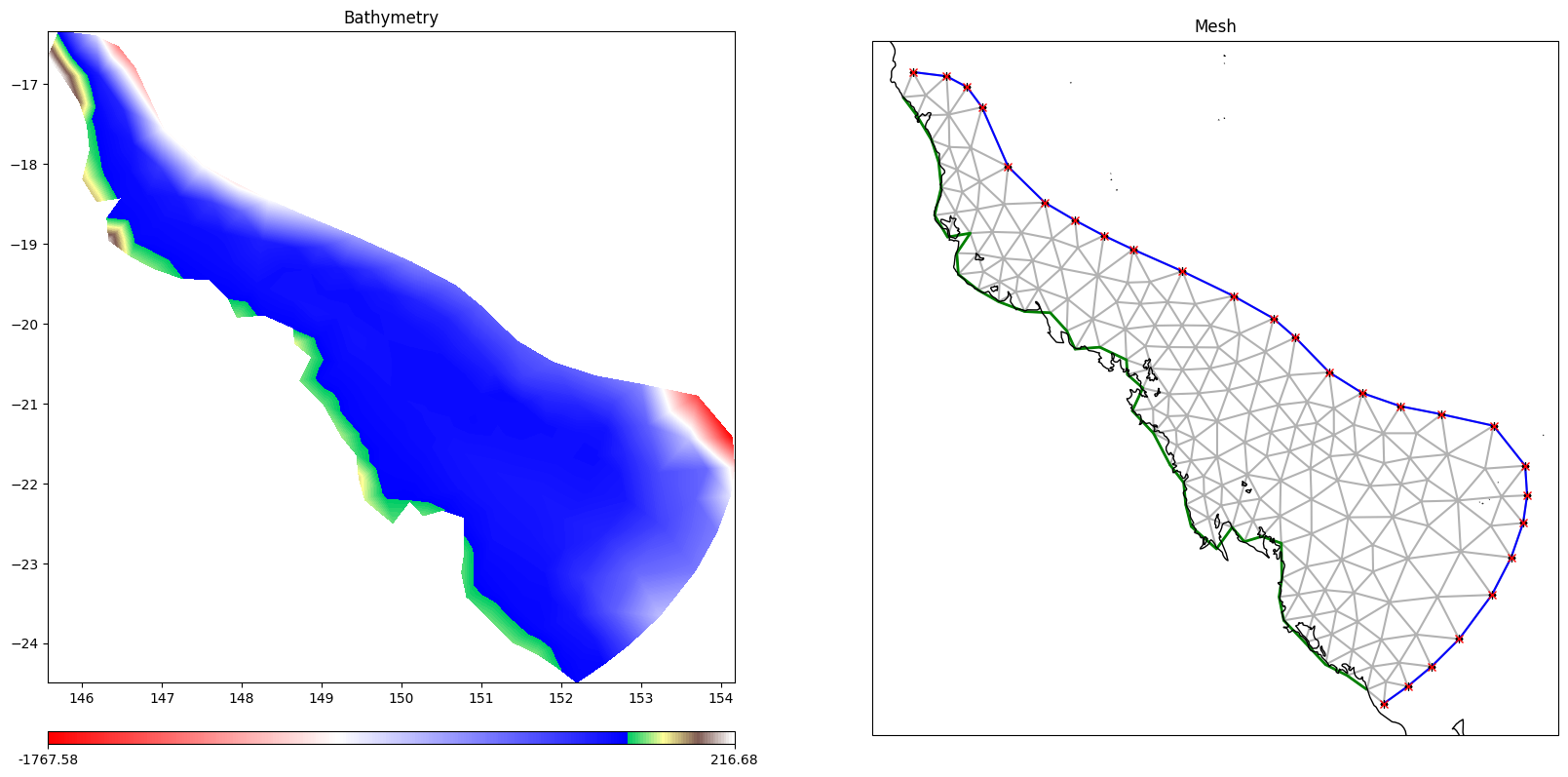
grid=SCHISMGrid(
hgrid=DataBlob(id="hgrid", source=hgrid),
drag=1,
)
grid.plot_hgrid()
WARNING:rompy.schism.grid:drag is being set to a constant value, this is not recommended. For best results, please supply friction gr3 files with spatially varying values. Further options are under development.
[5]:
grid.get(workdir)
list(workdir.glob('*'))
INFO:rompy.schism.grid:Generated albedo with constant value of 0.15
INFO:rompy.schism.grid:Generated diffmin with constant value of 1e-06
INFO:rompy.schism.grid:Generated diffmax with constant value of 1.0
INFO:rompy.schism.grid:Generated watertype with constant value of 1.0
INFO:rompy.schism.grid:Generated windrot_geo2proj with constant value of 0.0
INFO:rompy.schism.grid:Generated drag with constant value of 1.0
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_procedural/hgrid.ll
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_procedural/hgrid_WWM.gr3
[5]:
[PosixPath('schism_procedural/albedo.gr3'),
PosixPath('schism_procedural/hgrid_WWM.gr3'),
PosixPath('schism_procedural/vgrid.in'),
PosixPath('schism_procedural/diffmax.gr3'),
PosixPath('schism_procedural/hgrid.gr3'),
PosixPath('schism_procedural/wwmbnd.gr3'),
PosixPath('schism_procedural/diffmin.gr3'),
PosixPath('schism_procedural/drag.gr3'),
PosixPath('schism_procedural/watertype.gr3'),
PosixPath('schism_procedural/windrot_geo2proj.gr3'),
PosixPath('schism_procedural/hgrid.ll'),
PosixPath('schism_procedural/tvd.prop')]
Forcing data#
[6]:
# First lists import the main data classes
from rompy.schism.data import SCHISMDataSflux, SCHISMDataOcean, SCHISMDataWave, SCHISMDataTides
# Sets also import a few of the minor classes that are used in the construction of these main classes for use in this demo
from rompy.schism.data import SfluxSource, TidalDataset, SfluxAir, SCHISMDataBoundary
# And also lets import some of the core data source objects. These are data input abstractions that work in exactly the same way as
# with the swan classes, and can be used interchangeably in each of the data classes depending on the data source. We will use a
# bit of a mix here for illustration purposes.
from rompy.core.data import DataBlob
from rompy.core.source import SourceFile, SourceDataset, SourceIntake, SourceDatamesh, SourceWavespectra
Sflux Data#
[7]:
from rompy.schism.namelists import Sflux_Inputs
# SCHISMDataSflux??
# SfluxSource??
# Sflux_Inputs??
[8]:
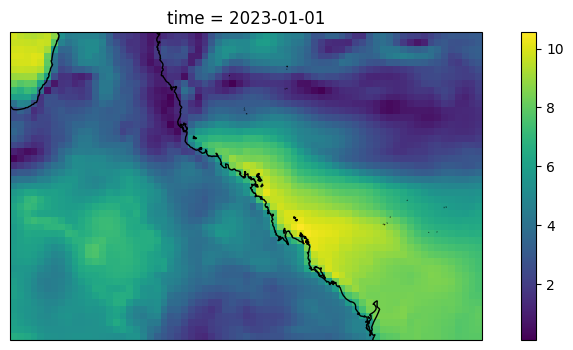
import intake
nn = 0
cat = intake.open_catalog(HERE / ".." / "data" / "catalog.yaml")
ds = xr.open_dataset(HERE/ "test_data" / "era5.nc")
plt.figure(figsize=(10,4))
ax = plt.axes(projection=ccrs.PlateCarree())
(np.sqrt(ds.u10[nn]**2 + ds.v10[nn]**2)).plot(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines()
[8]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x112170c71a60>
[9]:
ds
[9]:
<xarray.Dataset> Size: 3MB
Dimensions: (time: 73, latitude: 45, longitude: 69)
Coordinates:
* latitude (latitude) float32 180B -15.0 -15.25 -15.5 ... -25.5 -25.75 -26.0
* longitude (longitude) float32 276B 140.0 140.2 140.5 ... 156.5 156.8 157.0
* time (time) datetime64[ns] 584B 2023-01-01 ... 2023-01-04
Data variables:
msl (time, latitude, longitude) float32 907kB ...
u10 (time, latitude, longitude) float32 907kB ...
v10 (time, latitude, longitude) float32 907kB ...
Attributes:
Conventions: CF-1.7
GRIB_centre: ecmf
GRIB_centreDescription: European Centre for Medium-Range Weather Forecasts
GRIB_subCentre: 0
history: 2024-11-10T00:18 GRIB to CDM+CF via cfgrib-0.9.1...
institution: European Centre for Medium-Range Weather Forecasts
_coordinates: {"t":"time","x":"longitude","y":"latitude"}
_request_size: 907700
_domain_size: 226665[10]:
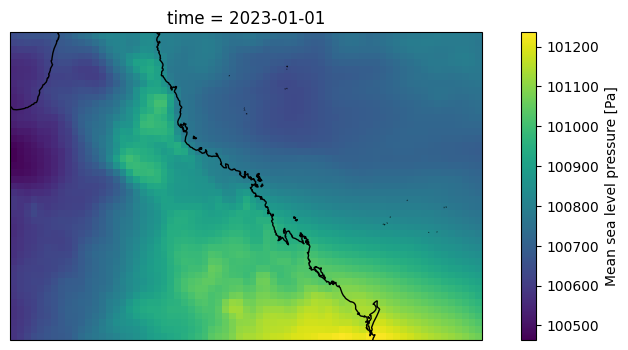
plt.figure(figsize=(10,4))
ax = plt.axes(projection=ccrs.PlateCarree())
ds.msl[nn].plot(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines()
[10]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x112168d287d0>
[11]:
# Lets have a look at an flux object. Here we will use a ERA5 dataset exposed through the intake catalog in the tests/data folder.
from rompy.core.time import TimeRange
atmos_forcing = SCHISMDataSflux(
air_1=SfluxAir(
id="air_1",
source=SourceFile(
uri=HERE / "test_data" / "era5.nc",
),
uwind_name='u10',
vwind_name='v10',
prmsl_name='msl',
filter={
"sort": {"coords": ["latitude"]},
},
buffer=2
)
)
atmos_forcing.get(destdir=workdir, grid=grid, time=TimeRange(start="2023-01-01", end="2023-01-02", dt=3600))
INFO:rompy.schism.data:Fetching air_1
[12]:
# Lets check the workdir again, we should see some prepared sflux inputs
list(workdir.glob('**/*'))
[12]:
[PosixPath('schism_procedural/sflux'),
PosixPath('schism_procedural/albedo.gr3'),
PosixPath('schism_procedural/hgrid_WWM.gr3'),
PosixPath('schism_procedural/vgrid.in'),
PosixPath('schism_procedural/diffmax.gr3'),
PosixPath('schism_procedural/hgrid.gr3'),
PosixPath('schism_procedural/wwmbnd.gr3'),
PosixPath('schism_procedural/diffmin.gr3'),
PosixPath('schism_procedural/drag.gr3'),
PosixPath('schism_procedural/watertype.gr3'),
PosixPath('schism_procedural/windrot_geo2proj.gr3'),
PosixPath('schism_procedural/hgrid.ll'),
PosixPath('schism_procedural/tvd.prop'),
PosixPath('schism_procedural/sflux/air_1.0001.nc'),
PosixPath('schism_procedural/sflux/sflux_inputs.txt')]
[13]:
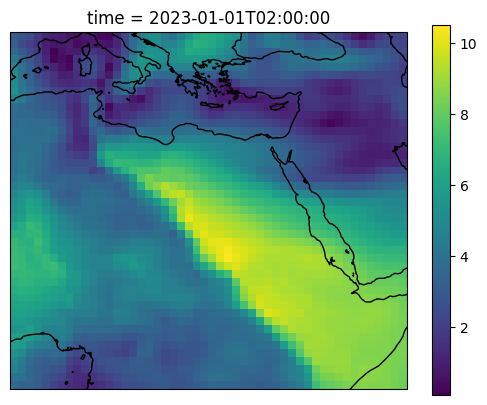
# Create a map
ax = plt.axes(projection=ccrs.PlateCarree())
# load the data
ds = xr.open_dataset("schism_procedural/sflux/air_1.0001.nc")
wind_speed = np.sqrt(ds.u10**2 + ds.v10**2)
# plot the data
wind_speed.isel(time=2).plot(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines()
[13]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x112168da5eb0>
Ocean Boundary#
[14]:
#SCHISMDataOcean??
[15]:
ds = xr.open_dataset(HERE / "test_data" / "hycom.nc")
ax = plt.axes(projection=ccrs.PlateCarree())
ds["surf_el"].isel(time=0).plot(ax=ax, transform=ccrs.PlateCarree())
ax.coastlines()
[15]:
<cartopy.mpl.feature_artist.FeatureArtist at 0x112168793f20>
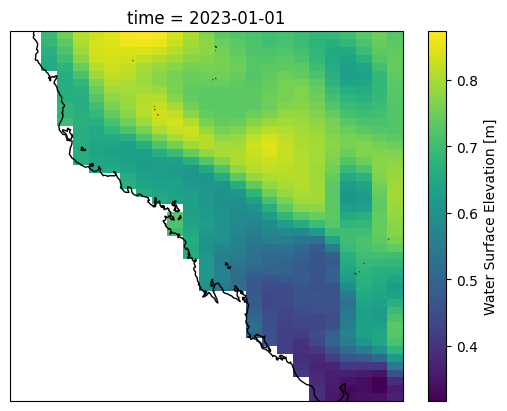
[16]:
ocean_forcing = SCHISMDataOcean(
elev2D = SCHISMDataBoundary(
id="hycom",
source=SourceFile(
uri=HERE / "test_data" / "hycom.nc",
),
variable="surf_el",
coords={"t": "time", "y": "ylat", "x": "xlon", "z": "depth"},
interpolate_missing_coastal=True,
),
)
[17]:
ocean_forcing.get(destdir=workdir, grid=grid)
list(workdir.glob("*"))
INFO:rompy.schism.data:Fetching elev2D
[17]:
[PosixPath('schism_procedural/sflux'),
PosixPath('schism_procedural/albedo.gr3'),
PosixPath('schism_procedural/hgrid_WWM.gr3'),
PosixPath('schism_procedural/vgrid.in'),
PosixPath('schism_procedural/diffmax.gr3'),
PosixPath('schism_procedural/hgrid.gr3'),
PosixPath('schism_procedural/wwmbnd.gr3'),
PosixPath('schism_procedural/diffmin.gr3'),
PosixPath('schism_procedural/drag.gr3'),
PosixPath('schism_procedural/watertype.gr3'),
PosixPath('schism_procedural/windrot_geo2proj.gr3'),
PosixPath('schism_procedural/hgrid.ll'),
PosixPath('schism_procedural/elev2D.th.nc'),
PosixPath('schism_procedural/tvd.prop')]
[18]:
dsb = xr.open_dataset('schism_procedural/elev2D.th.nc')
dsb.time[0]
[18]:
<xarray.DataArray 'time' ()> Size: 8B
array('2023-01-01T00:00:00.000000000', dtype='datetime64[ns]')
Coordinates:
time datetime64[ns] 8B 2023-01-01
Attributes:
long_name: Time
standard_name: time
base_date: [2023 1 1 0 0 0][19]:
vmin, vmax = 0.3, 0.9
time = dsb.time[0]
ax = plt.axes(projection=ccrs.PlateCarree())
ds["surf_el"].sel(time=time).plot(ax=ax, transform=ccrs.PlateCarree(), vmin=vmin, vmax=vmax)
ax.coastlines()
values = dsb.time_series.isel(time=0)
x,y = grid.boundary_points()
ax.scatter(x, y, transform=ccrs.PlateCarree(), c=values, cmap="viridis", vmin=vmin, vmax=vmax, edgecolor="black")
# Check for nans (there shouldn't be any)
# nans = dsb.time_series.isel(time=0).isnull().squeeze()
# ax.scatter(x[nans], y[nans], transform=ccrs.PlateCarree(), c="red", edgecolor="black")
[19]:
<matplotlib.collections.PathCollection at 0x112170c71f70>
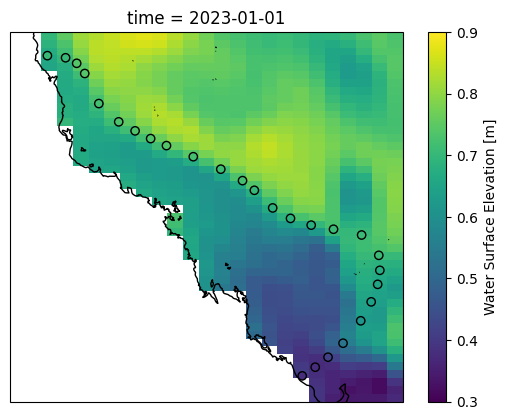
[20]:
ocean_forcing.elev2D.ds
[20]:
<xarray.Dataset> Size: 10kB
Dimensions: (time: 2, ylat: 47, xlon: 25)
Coordinates:
* ylat (ylat) float64 376B -25.04 -24.84 -24.64 ... -16.24 -16.04 -15.84
* xlon (xlon) float64 200B 145.0 145.4 145.8 146.2 ... 153.8 154.2 154.6
* time (time) datetime64[ns] 16B 2023-01-01 2023-01-02
Data variables:
surf_el (time, ylat, xlon) float32 9kB ...
Attributes:
classification_level: UNCLASSIFIED
distribution_statement: Approved for public release. Distribution unli...
downgrade_date: not applicable
classification_authority: not applicable
institution: Fleet Numerical Meteorology and Oceanography C...
source: HYCOM archive file
history: Thu Dec 21 12:28:25 2023: ncrcat hycom_2023010...
comment: p-grid
field_type: instantaneous
Conventions: CF-1.6 NAVO_netcdf_v1.1
NCO: netCDF Operators version 5.1.8 (Homepage = htt...Wave#
[21]:
# SCHISMDataWave??
[22]:
wave_forcing = SCHISMDataWave(
id="wavedata",
source=SourceIntake(
dataset_id="ausspec",
catalog_uri=HERE / ".." / "data" / "catalog.yaml",
),
coords={'x': "lon", 'y': "lat"},
)
[23]:
ax = wave_forcing.plot(model_grid=grid)
wave_forcing.plot_boundary(ax=ax, grid=grid)
[23]:
<GeoAxes: >
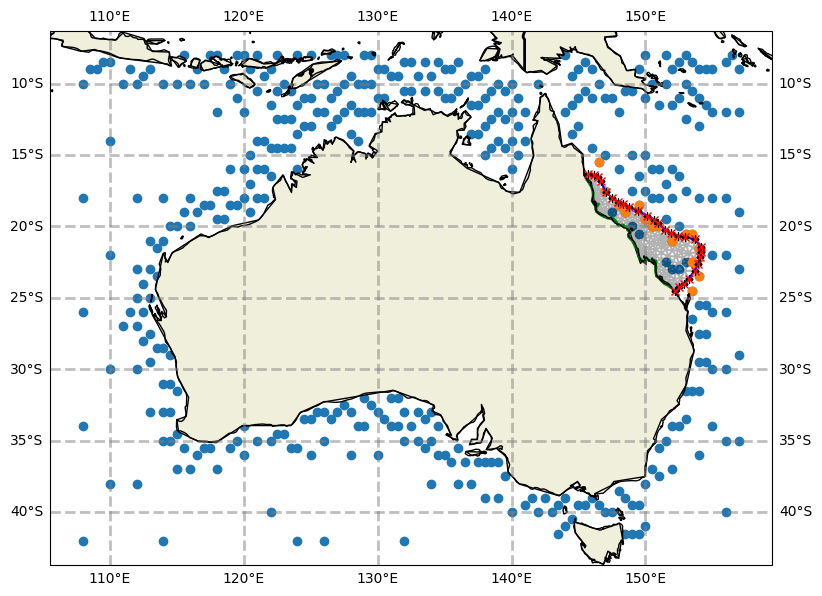
Tidal data#
[24]:
# SCHISMDataTides?
# TidalDataset?
[25]:
tidal_forcing = SCHISMDataTides(
tidal_data=TidalDataset(
elevations=HERE / "test_data"/ "tpxo9-neaus" / "h_m2s2n2.nc",
velocities=HERE / "test_data"/ "tpxo9-neaus" / "u_m2s2n2.nc"
),
constituents=["M2", "S2", "N2"],
cutoff_depth=50,
flags=[[5, 3, 0, 0]]
)
tidal_forcing.get(
destdir=workdir,
grid=grid,
time=TimeRange(start="2023-01-01", end="2023-01-02", dt=3600),
)
INFO:rompy.schism.data:Generating tides
INFO:pyschism.forcing.bctides.bctides:Processing boundary 1:
INFO:pyschism.forcing.bctides.bctides:Elevation type: 5
WARNING:pyschism.forcing.bctides.bctides:Combination of 3 and 4, time history of elevation is read in from elev2D.th.nc!
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent M2.
INFO:pyschism.forcing.bctides.tpxo:h_file is ../../tests/schism/test_data/tpxo9-neaus/h_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent N2.
INFO:pyschism.forcing.bctides.bctides:Velocity type: 3
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent M2.
INFO:pyschism.forcing.bctides.tpxo:u_file is ../../tests/schism/test_data/tpxo9-neaus/u_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent N2.
INFO:pyschism.forcing.bctides.bctides:Temperature type: 0
WARNING:pyschism.forcing.bctides.bctides:Temperature is not sepcified, not input needed!
INFO:pyschism.forcing.bctides.bctides:Salinity type: 0
INFO:pyschism.forcing.bctides.bctides:Salinity is not sepcified, not input needed!
Full config object#
[26]:
# Instantiate a config object
from rompy.schism import SchismCSIROConfig
from rompy.schism.data import SCHISMData
from pydantic import ValidationError
try:
config=SchismCSIROConfig(
grid=grid,
data=SCHISMData(
atmos=atmos_forcing,
ocean=ocean_forcing,
wave=wave_forcing,
tides=tidal_forcing
),
)
except ValidationError as e:
print(e)
1 validation error for SchismCSIROConfig
Value error, manning.gr3 must be specified when nchi=-1 [type=value_error, input_value={'grid': SCHISMGrid([152....], sobc=[1], relax=[]))}, input_type=dict]
For further information visit https://errors.pydantic.dev/2.10/v/value_error
[27]:
# That gives us an expected error due to teh fact that we have a validator checking required inputs
# Lets fix the grid issue and try again
try:
config=SchismCSIROConfig(
grid=grid,
data=SCHISMData(
atmos=atmos_forcing,
ocean=ocean_forcing,
wave=wave_forcing,
tides=tidal_forcing
),
)
except ValidationError as e:
print(e)
# Again we get a validation error, the hgrid_WWM is missing. Lets add it and try again
grid=SCHISMGrid(
hgrid=DataBlob(id="hgrid", source=hgrid),
manning=1,
)
config=SchismCSIROConfig(
grid=grid,
mesbf=1,
fricc=0.067,
param_iof_hydro1=1, # elevation
param_iof_hydro14=1, # wind speed
param_iof_hydro16=1, # surface velocities
wwm18=1, # peak wave direction
wwm1=1, # significant wave height
wwm9=1, # peak period
data=SCHISMData(
atmos=atmos_forcing,
ocean=ocean_forcing,
wave=wave_forcing,
tides=tidal_forcing
),
)
WARNING:rompy.schism.grid:manning is being set to a constant value, this is not recommended. For best results, please supply friction gr3 files with spatially varying values. Further options are under development.
1 validation error for SchismCSIROConfig
Value error, manning.gr3 must be specified when nchi=-1 [type=value_error, input_value={'grid': SCHISMGrid([152....], sobc=[1], relax=[]))}, input_type=dict]
For further information visit https://errors.pydantic.dev/2.10/v/value_error
Model Run#
Note that most fields are optional, this sample using defaults values.
Generate workspace#
[28]:
#if workdir.exists():
# rmtree(workdir)
#workdir.mkdir(exist_ok=True)
from rompy.model import ModelRun
from rompy.schism import SchismCSIROConfig
run = ModelRun(
run_id="test_schism",
period=TimeRange(start=datetime(2023, 1, 1, 0), end=datetime(2023, 1, 1, 12), interval="1h"),
output_dir=str(workdir),
config=config
)
rundir = run()
INFO:rompy.model:
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Model settings:
INFO:rompy.model:
run_id: test_schism
period:
Start: 2023-01-01 00:00:00
End: 2023-01-01 12:00:00
Duration: 12:00:00
Interval: 1:00:00
Include End: True
output_dir: schism_procedural
config: <class 'rompy.schism.config.SchismCSIROConfig'>
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Generating model input files in schism_procedural
INFO:rompy.schism.grid:Generated albedo with constant value of 0.15
INFO:rompy.schism.grid:Generated diffmin with constant value of 1e-06
INFO:rompy.schism.grid:Generated diffmax with constant value of 1.0
INFO:rompy.schism.grid:Generated watertype with constant value of 1.0
INFO:rompy.schism.grid:Generated windrot_geo2proj with constant value of 0.0
INFO:rompy.schism.grid:Generated manning with constant value of 1.0
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_procedural/test_schism/hgrid.ll
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_procedural/test_schism/hgrid_WWM.gr3
INFO:rompy.schism.data:Fetching air_1
INFO:rompy.schism.data:Fetching elev2D
INFO:rompy.schism.data:Fetching wavedata
INFO:rompy.schism.data:Generating tides
INFO:pyschism.forcing.bctides.bctides:Processing boundary 1:
INFO:pyschism.forcing.bctides.bctides:Elevation type: 5
WARNING:pyschism.forcing.bctides.bctides:Combination of 3 and 4, time history of elevation is read in from elev2D.th.nc!
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent M2.
INFO:pyschism.forcing.bctides.tpxo:h_file is ../../tests/schism/test_data/tpxo9-neaus/h_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent N2.
INFO:pyschism.forcing.bctides.bctides:Velocity type: 3
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent M2.
INFO:pyschism.forcing.bctides.tpxo:u_file is ../../tests/schism/test_data/tpxo9-neaus/u_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent N2.
INFO:pyschism.forcing.bctides.bctides:Temperature type: 0
WARNING:pyschism.forcing.bctides.bctides:Temperature is not sepcified, not input needed!
INFO:pyschism.forcing.bctides.bctides:Salinity type: 0
INFO:pyschism.forcing.bctides.bctides:Salinity is not sepcified, not input needed!
INFO:rompy.model:
INFO:rompy.model:Successfully generated project in schism_procedural
INFO:rompy.model:-----------------------------------------------------
[29]:
#wave_forcing = DataBlob(id="wavedata", source="../../tests/data/wavedata.nc")
Run schism#
(Note that paths to schism binaries will have to be updated)
[30]:
# Run the model
# !cd schism_procedural/test_schism && mpirun -np 4 /source/schism/src/schism_VL_WWM_v59 1
# Combine outputs
# !cd schism_procedural/test_schism/outputs && /source/schism/src/Utility/Combining_Scripts/combine_output11.exe -b 1 -e 1
# Use docker
!docker run -v ./schism_procedural/test_schism:/tmp/schism schism mpiexec -np 8 --allow-run-as-root pschism_git_HYDRO_VL_WWM 1
!docker run -v ./schism_procedural/test_schism/outputs:/tmp/schism schism combine_output11 -b 1 -e 1
Begin: 1, End: 1, dry flag: 0
combine all variables
Check results#
[31]:
list(Path(f"{rundir}/outputs").glob("*"))
[31]:
[PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0003'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0000_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0007_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/readme.md'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0007'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0007'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0006_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/flux.out'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/coriolis.out'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0006'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0007'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0003'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0004_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0002_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0004'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0005'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/fatal.error'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/param.out.nml'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0004'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0001'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0003_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/total.out'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0006'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0005'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0000'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0000'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0001'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0002'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0005'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0002'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0001'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0005_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/mirror.out'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0004'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0002'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0000'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0003'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0000'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0003'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/global_to_local.prop'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/schout_0001_1.nc'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0005'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0001'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxdahv_0002'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/local_to_global_0006'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0007'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/total_TR.out'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/subcycling.out'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/nonfatal_0006'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/maxelev_0004'),
PosixPath('/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_procedural/test_schism/outputs/JCG.out')]
[32]:
#load schism files
from rompy.schism.utils import schism_load, schism_plot
schfile=('schism_procedural/test_schism/outputs/schout_1.nc')
schout,meshtri=schism_load(schfile)
lons = schout.SCHISM_hgrid_node_y.values
lats = schout.SCHISM_hgrid_node_x.values
schout
[32]:
<xarray.Dataset> Size: 207kB
Dimensions: (time: 12, one: 1, nSCHISM_hgrid_face: 317,
nMaxSCHISM_hgrid_face_nodes: 4,
nSCHISM_hgrid_edge: 508, two: 2,
nSCHISM_hgrid_node: 192, sigma: 2,
nSCHISM_vgrid_layers: 2)
Coordinates:
* time (time) datetime64[ns] 96B 2023-01-01T01:00:00 .....
* sigma (sigma) float32 8B -1.0 0.0
Dimensions without coordinates: one, nSCHISM_hgrid_face,
nMaxSCHISM_hgrid_face_nodes,
nSCHISM_hgrid_edge, two, nSCHISM_hgrid_node,
nSCHISM_vgrid_layers
Data variables: (12/35)
SCHISM_hgrid (one) int32 4B ...
SCHISM_hgrid_face_nodes (nSCHISM_hgrid_face, nMaxSCHISM_hgrid_face_nodes) float64 10kB ...
SCHISM_hgrid_edge_nodes (nSCHISM_hgrid_edge, two) int32 4kB ...
SCHISM_hgrid_node_x (nSCHISM_hgrid_node) float64 2kB 152.2 ... 145.6
SCHISM_hgrid_node_y (nSCHISM_hgrid_node) float64 2kB -24.49 ... -16.67
node_bottom_index (nSCHISM_hgrid_node) int32 768B ...
... ...
WWM_2 (time, nSCHISM_hgrid_node) float32 9kB ...
WWM_10 (time, nSCHISM_hgrid_node) float32 9kB ...
WWM_11 (time, nSCHISM_hgrid_node) float32 9kB ...
WWM_18 (time, nSCHISM_hgrid_node) float32 9kB ...
WWM_19 (time, nSCHISM_hgrid_node) float32 9kB ...
WWM_20 (time, nSCHISM_hgrid_node) float32 9kB ...
Attributes:
Conventions: CF-1.0, UGRID-1.0
title: SCHISM Model output
institution: SCHISM Model output
source: SCHISM model output version v10
references: http://ccrm.vims.edu/schismweb/
history: created by combine_output11
comment: SCHISM Model output
type: SCHISM Model output
VisIT_plugin: https://schism.water.ca.gov/library/-/document_library/vie...[33]:
# plot gridded fields - elevation
for ix, time in enumerate(schout.time.values):
fig, ax =schism_plot(schout, meshtri,'elev', bbox=[145,-25,155,-16], project=True, plotmesh=True, mask=False, time=time,
vectors=True, contours=[0])
ax.set_title(time)
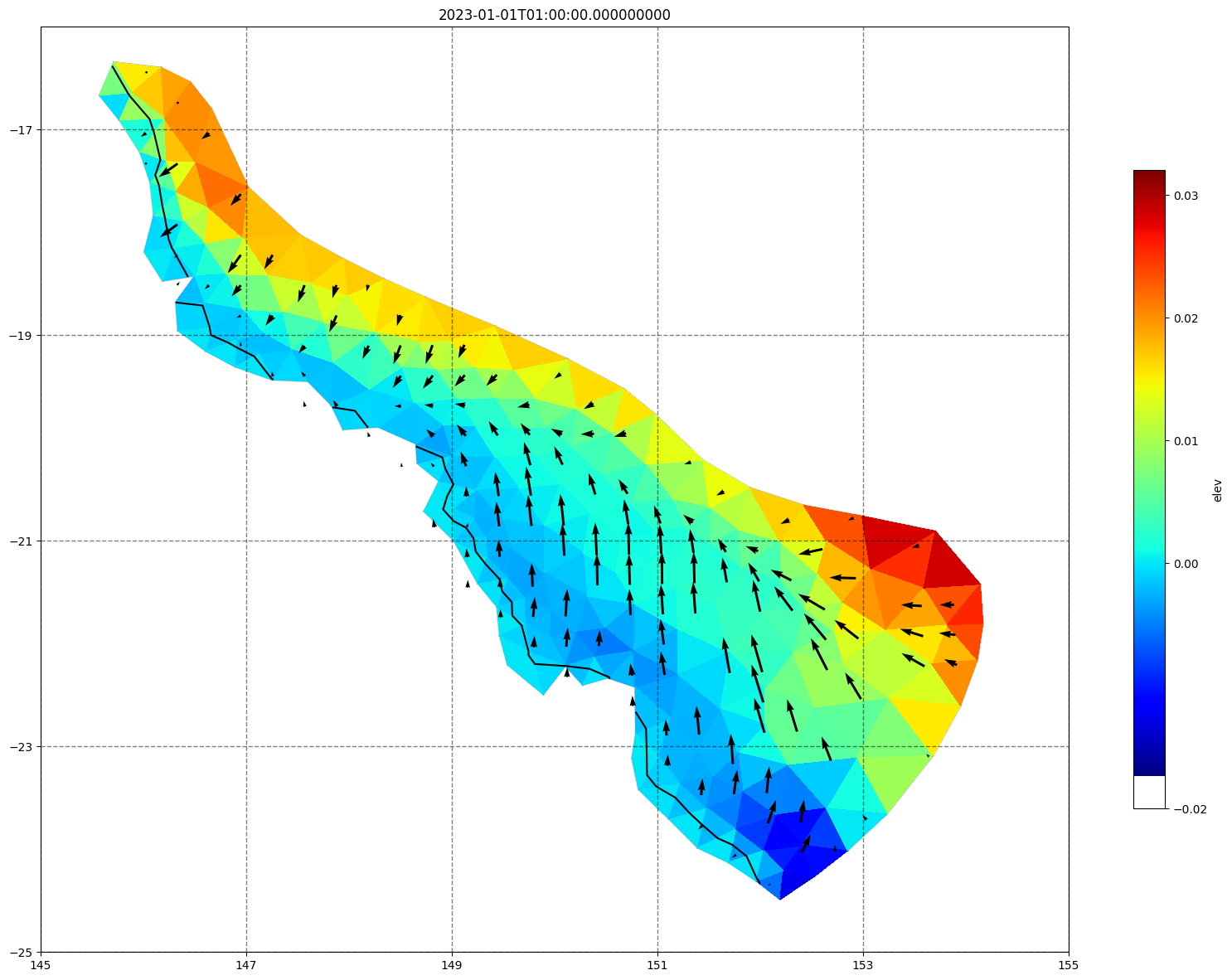
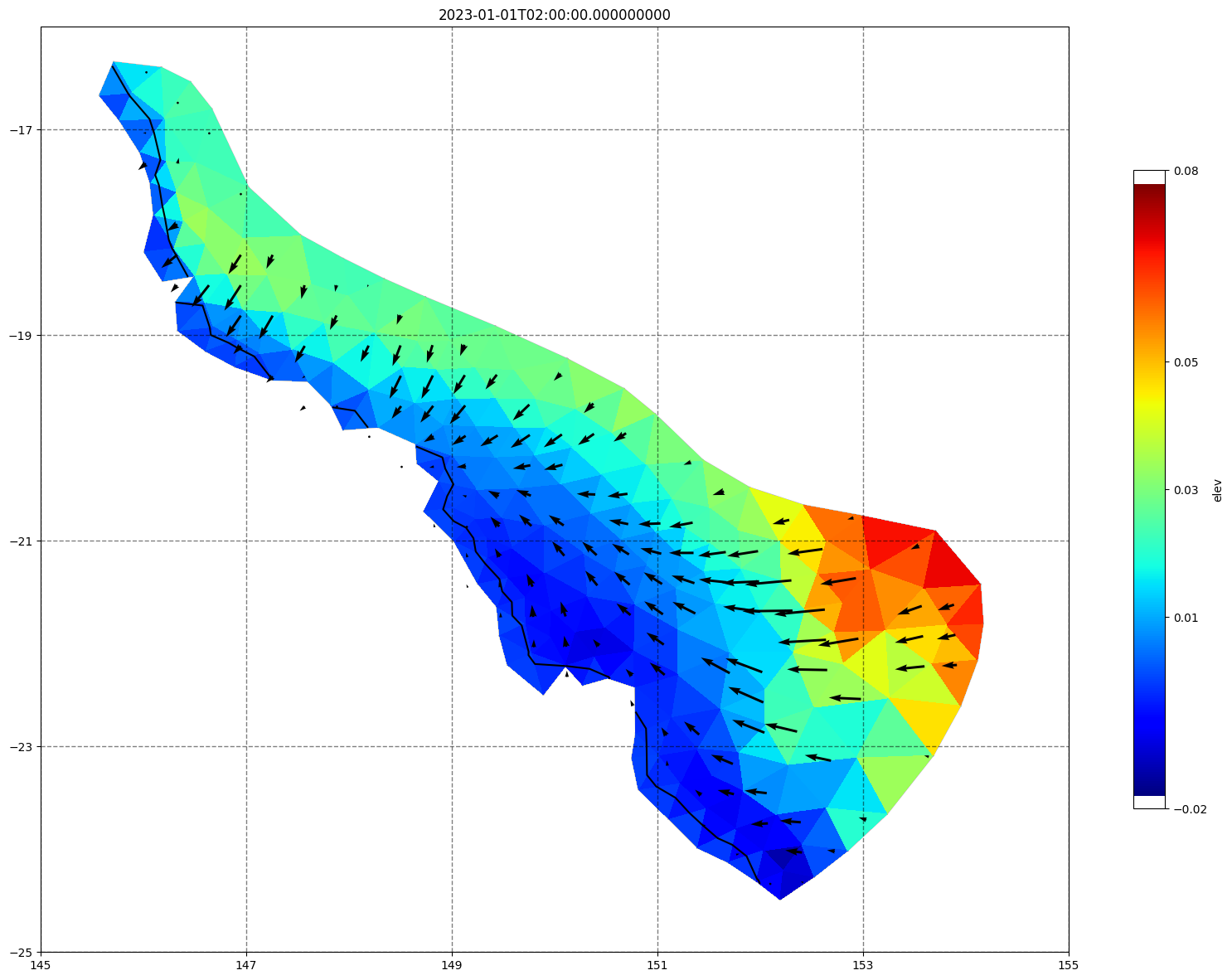
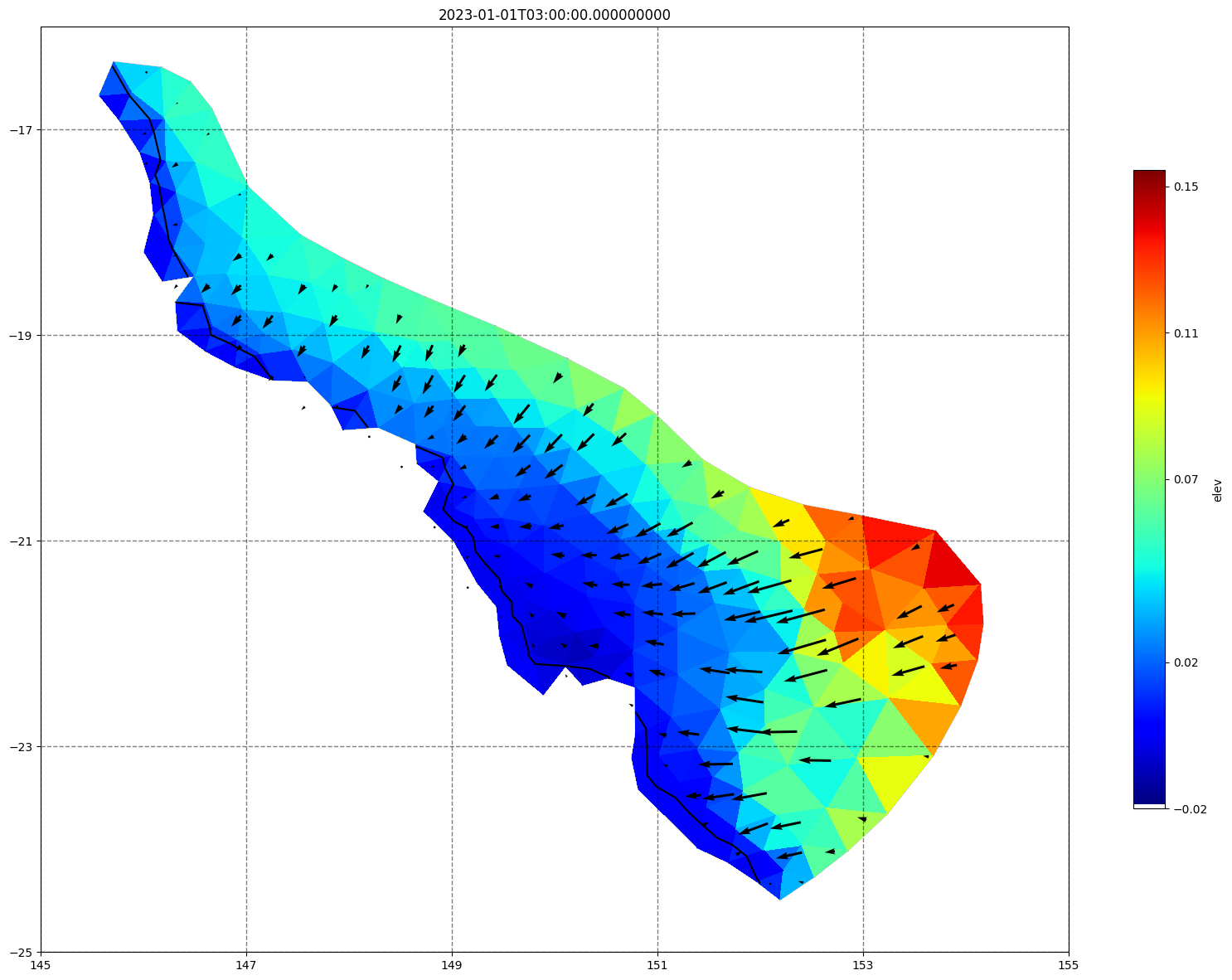

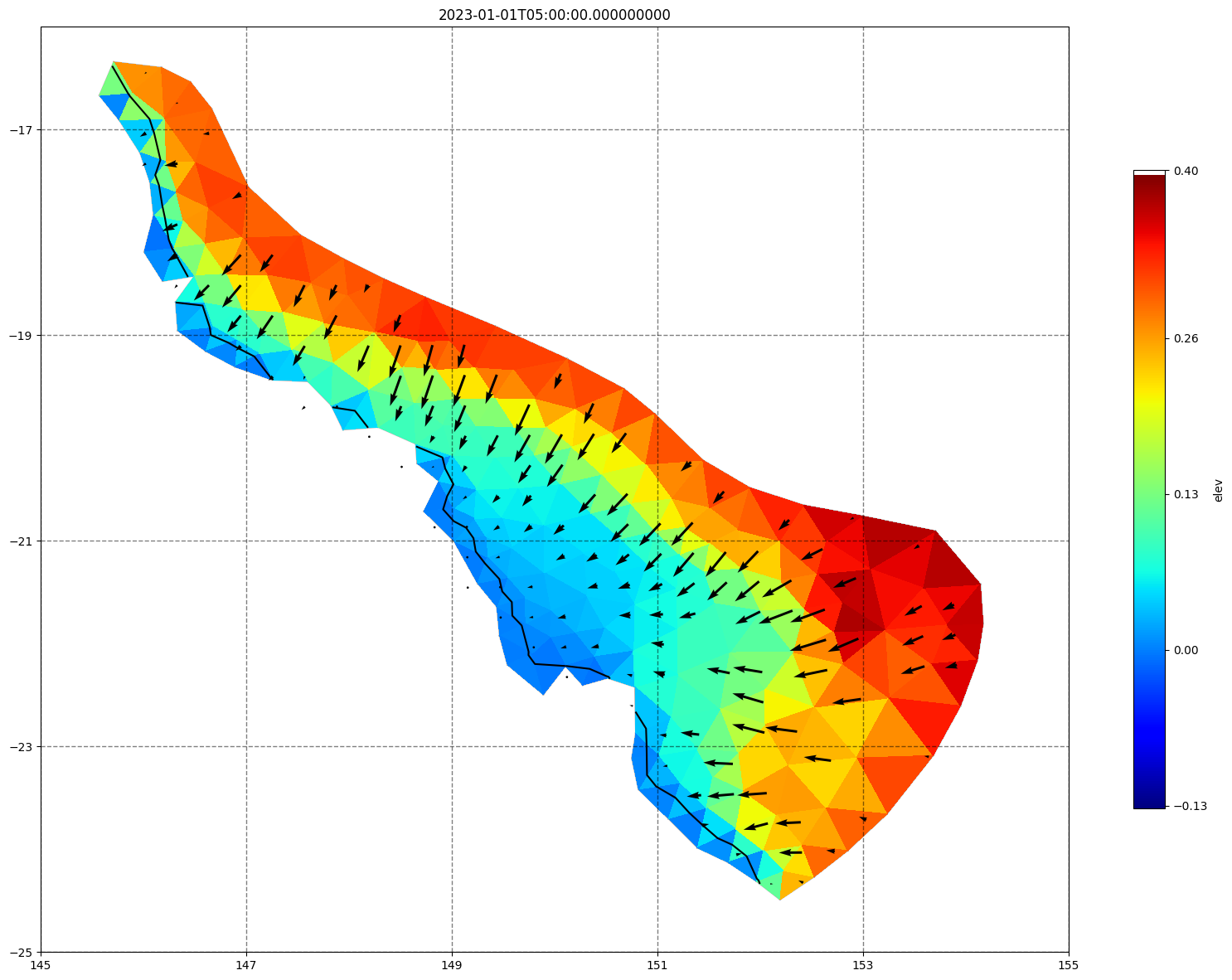
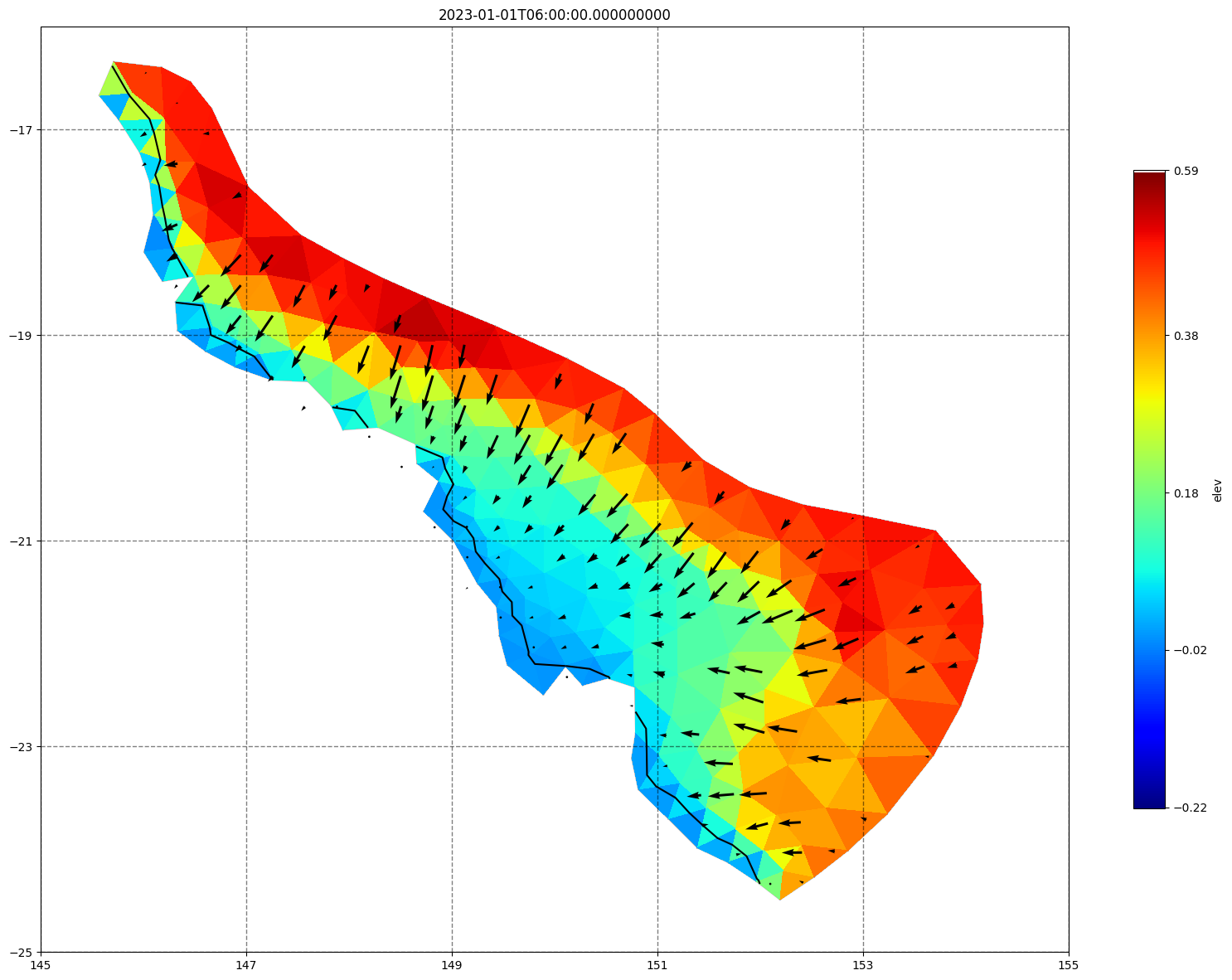
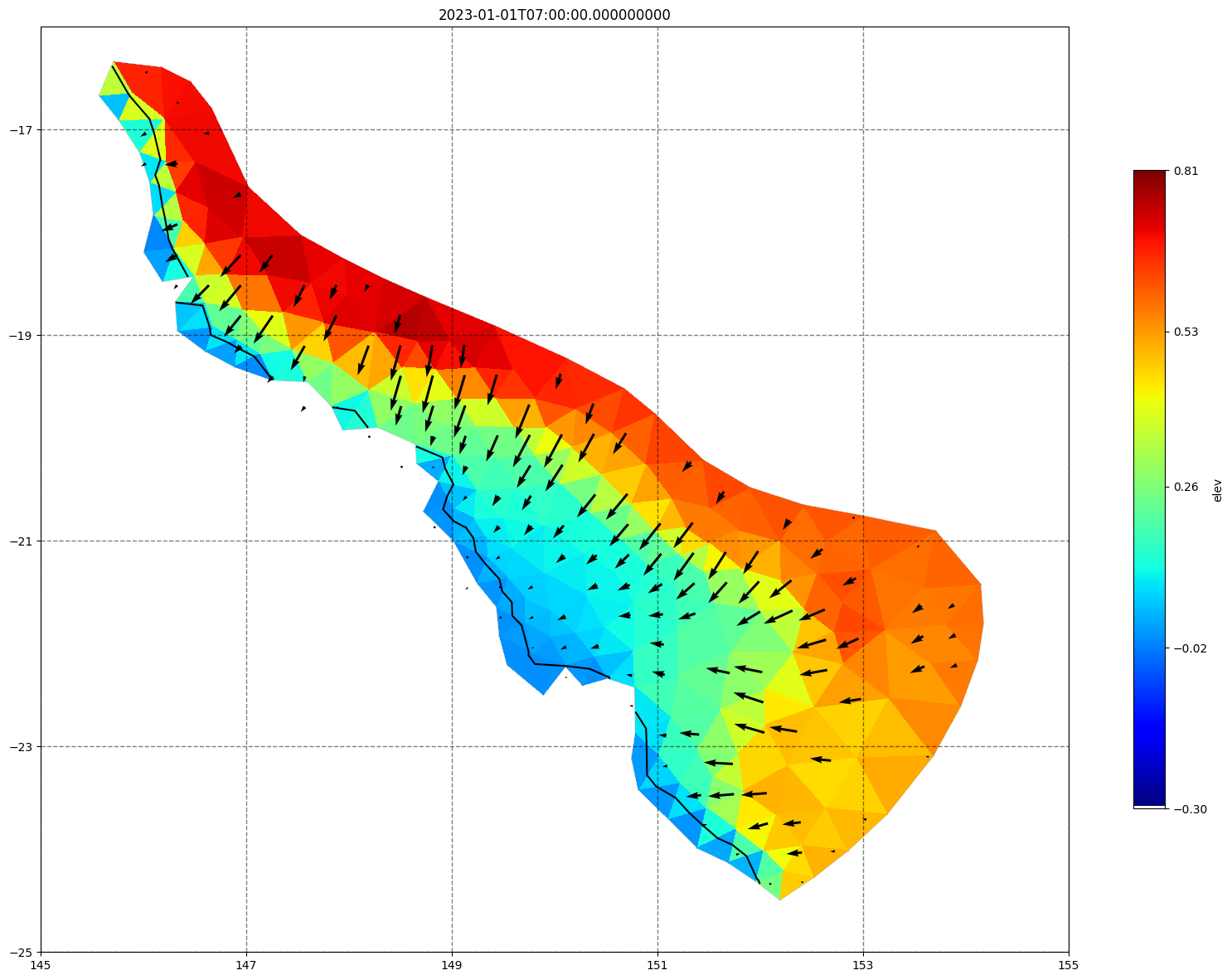
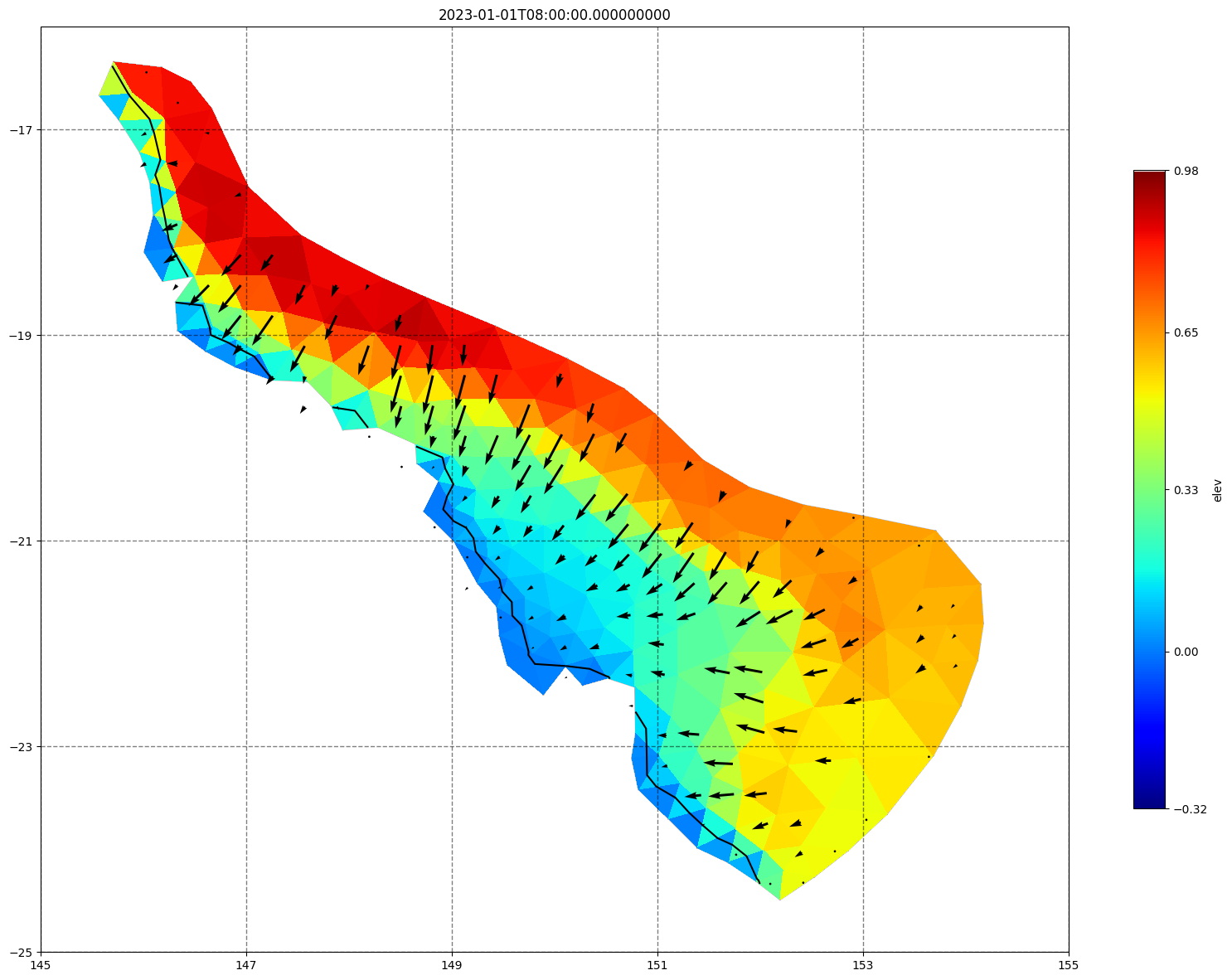
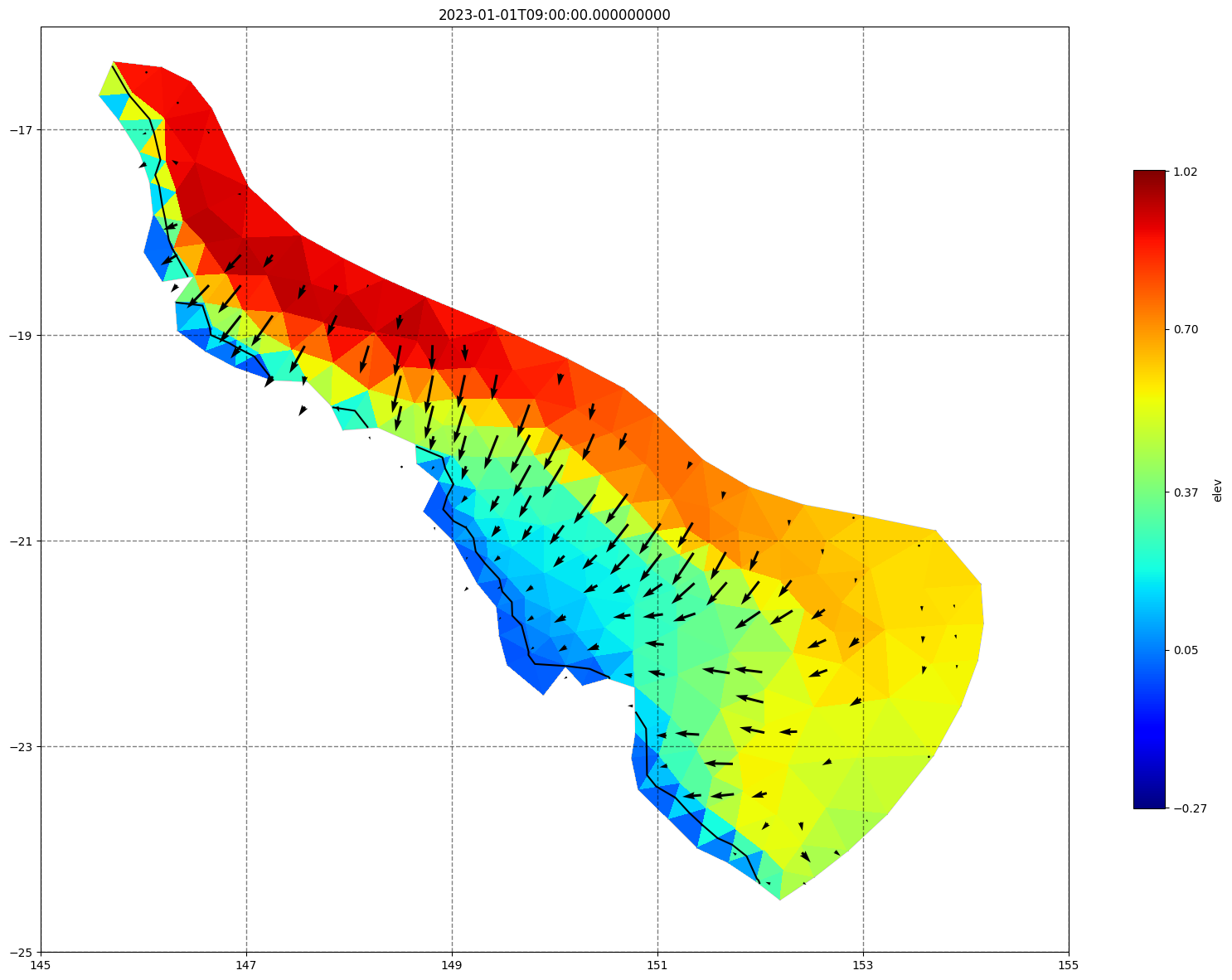
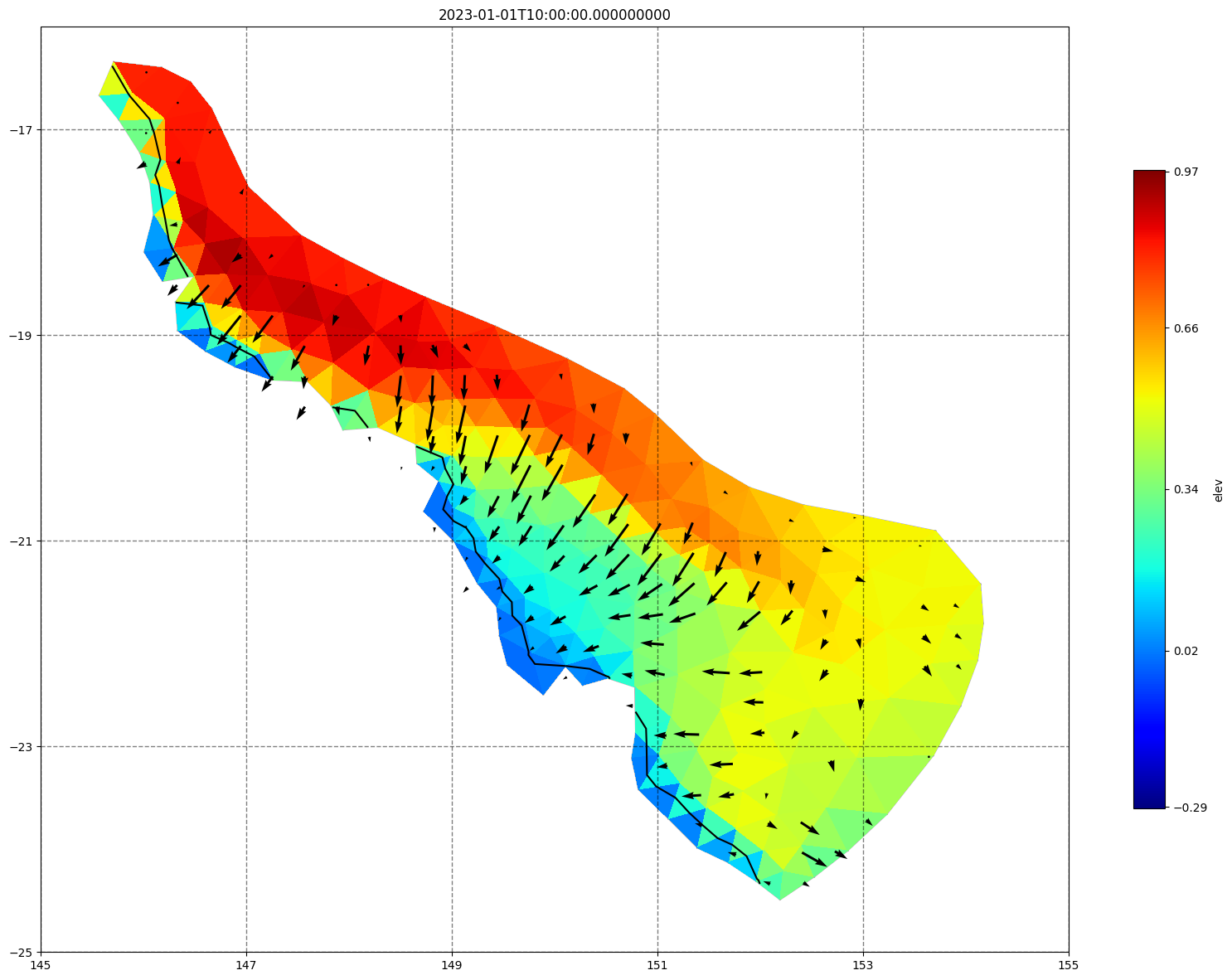
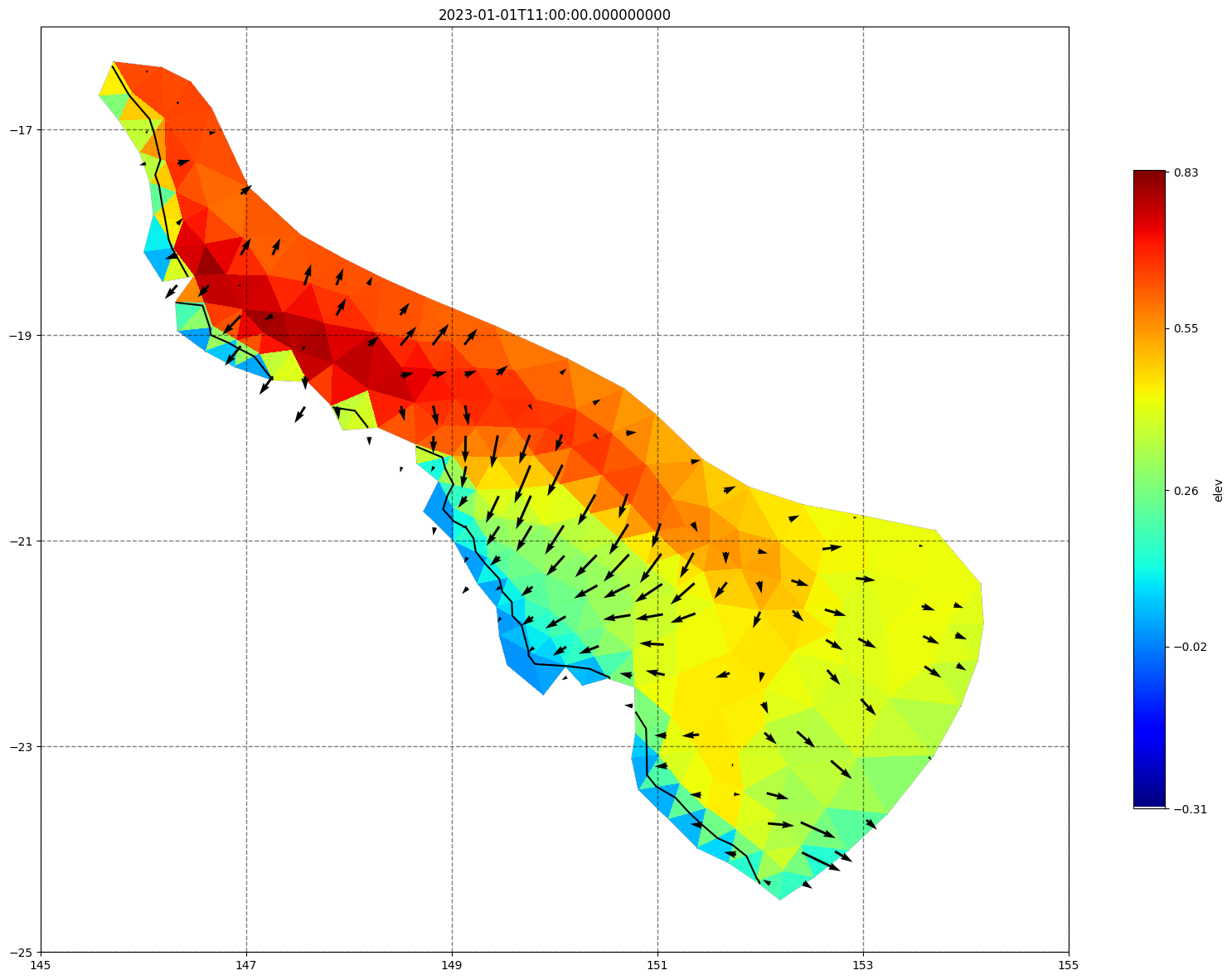
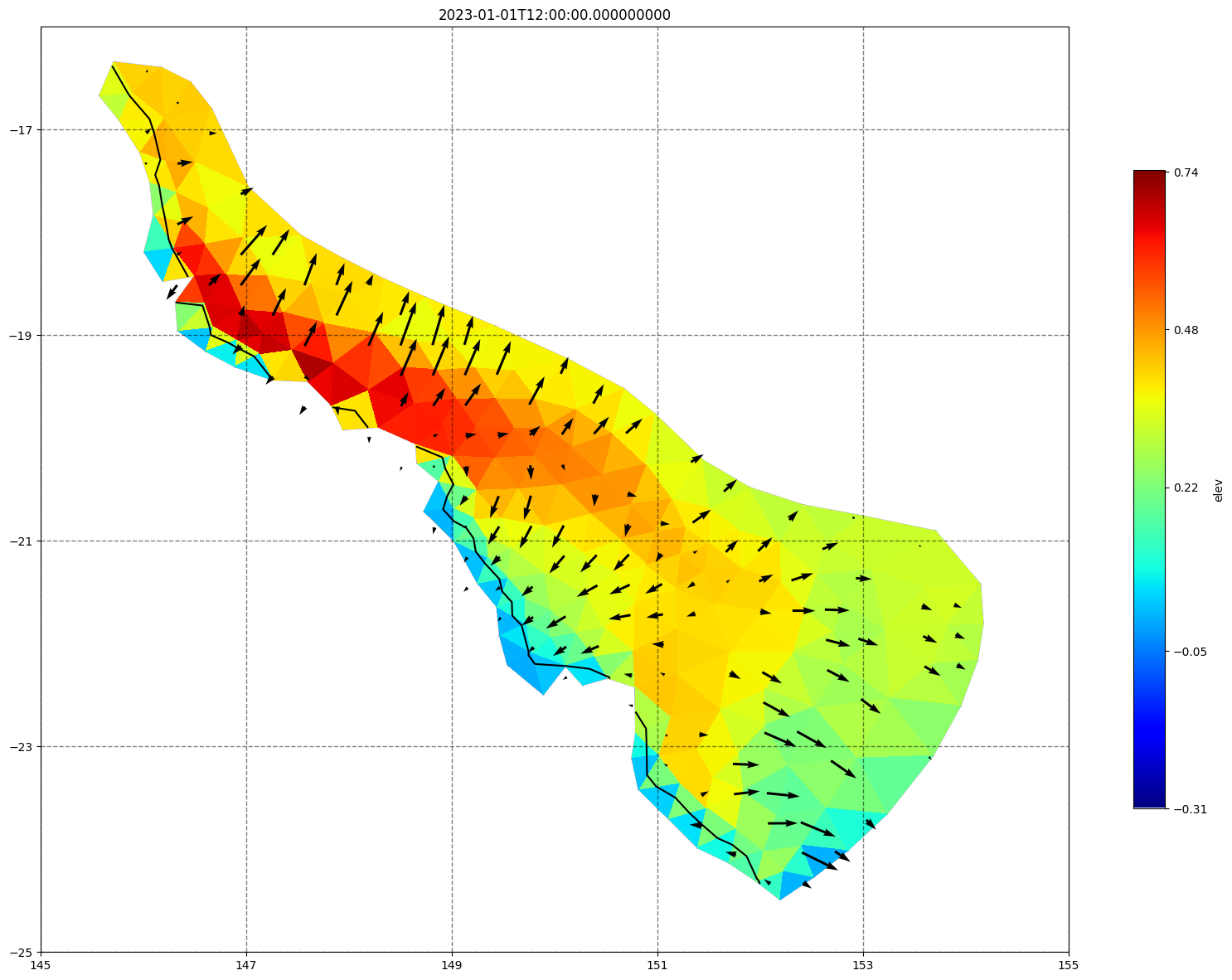
[34]:
## plot gridded fields - Hs
for ix, time in enumerate(schout.time.values):
fig, ax=schism_plot(schout, meshtri,'WWM_1', bbox=[145,-25,155,-16], project=True, plotmesh=True, mask=False, time=time,
vectors=True, contours=[0])
ax.set_title(time)
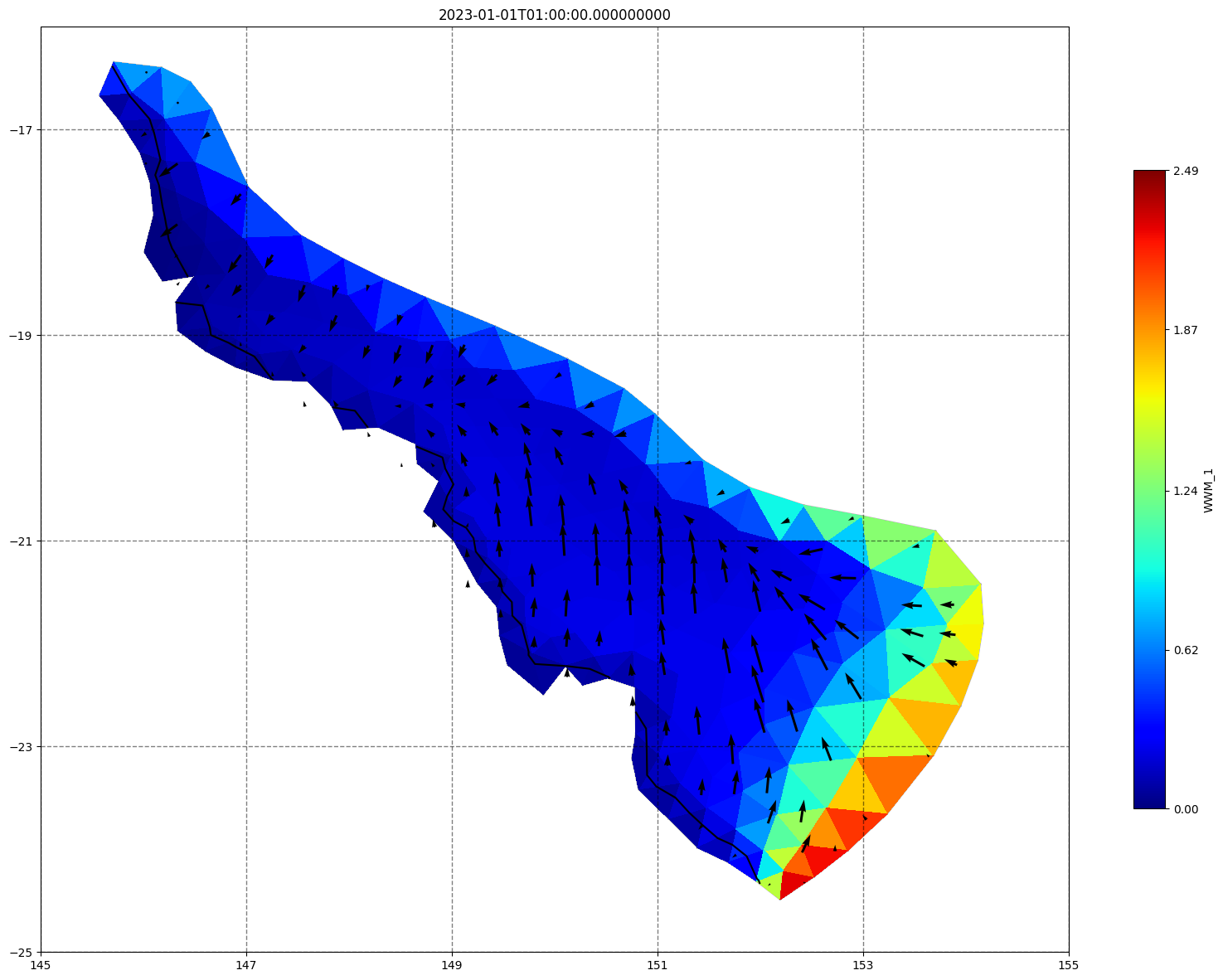
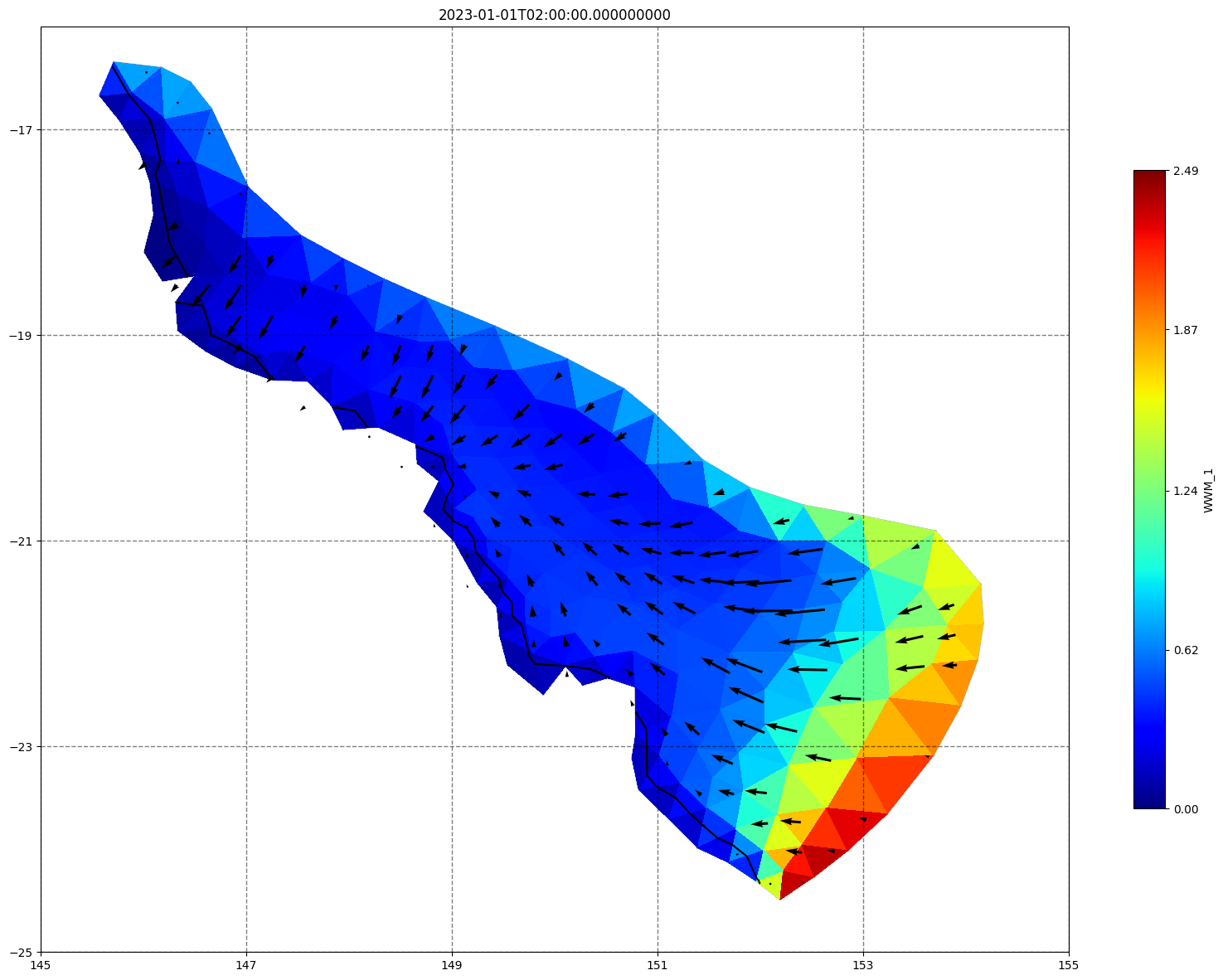
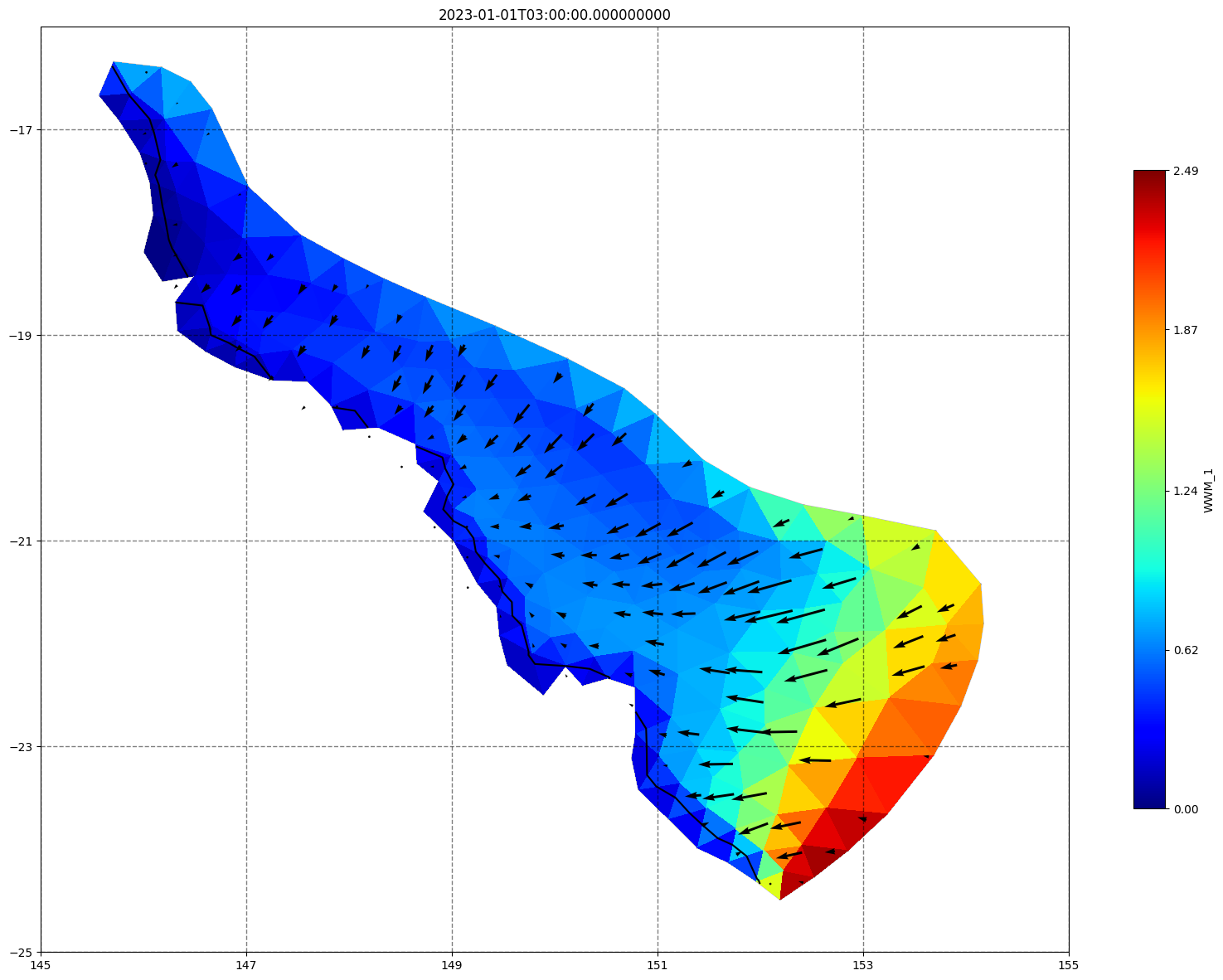
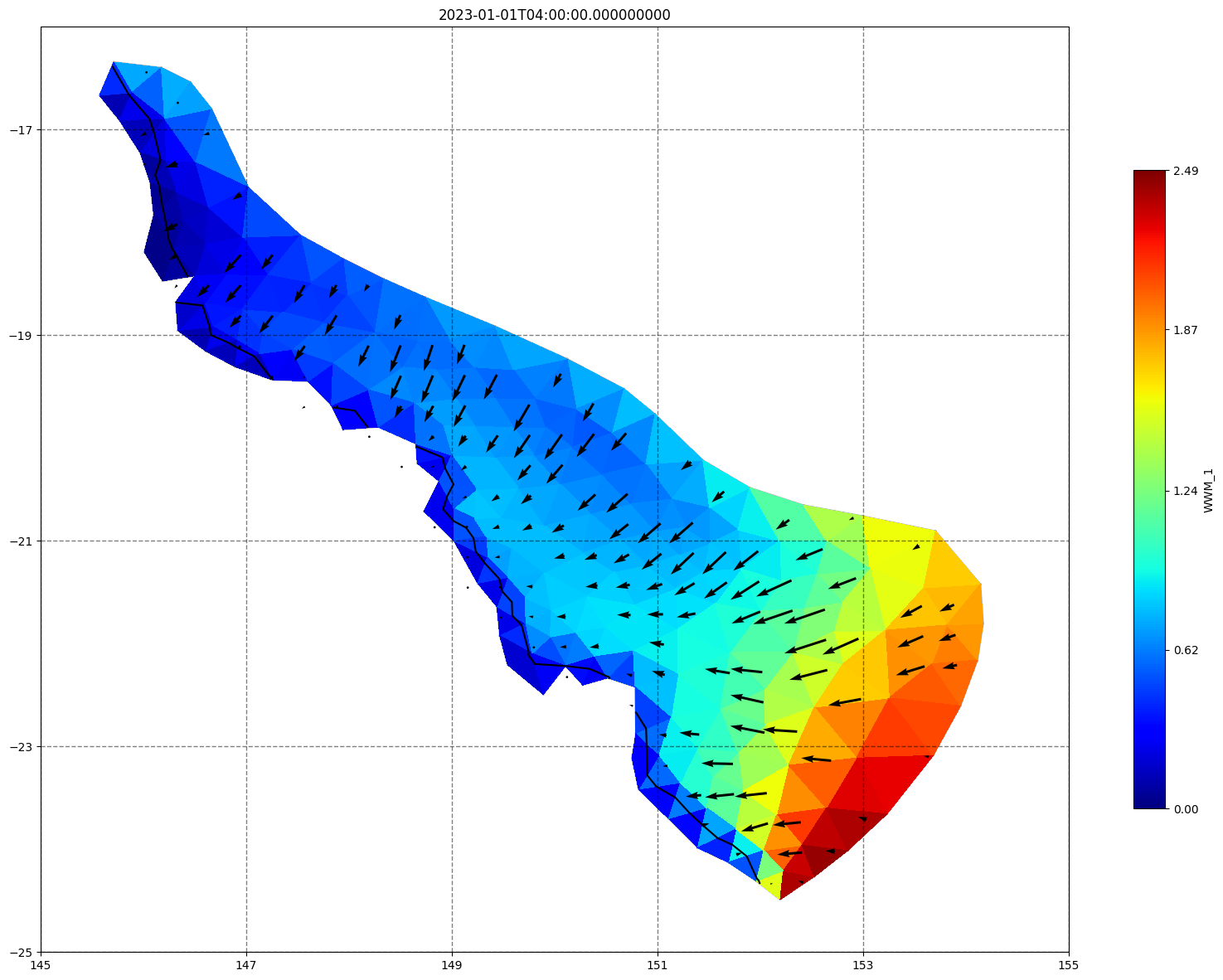
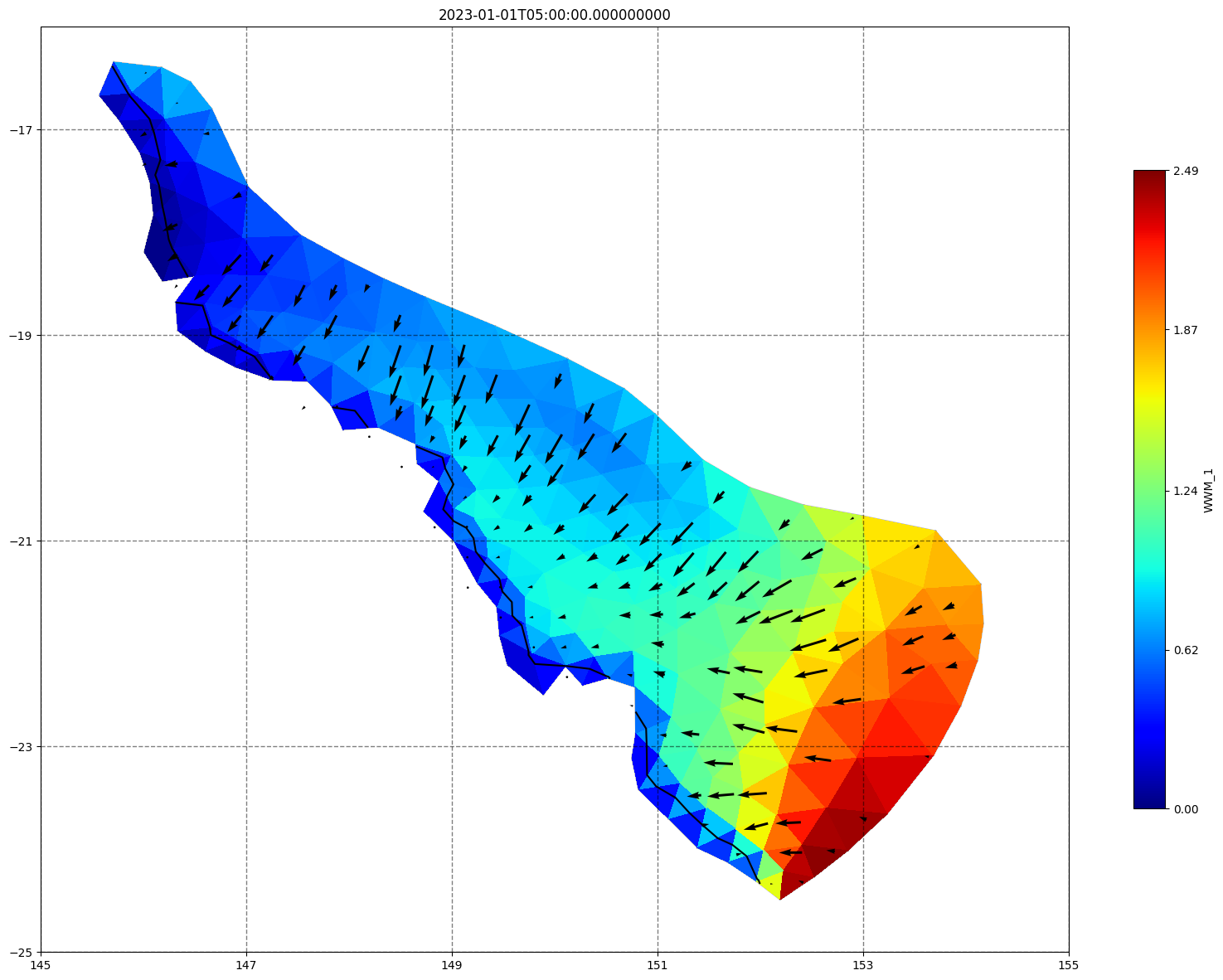
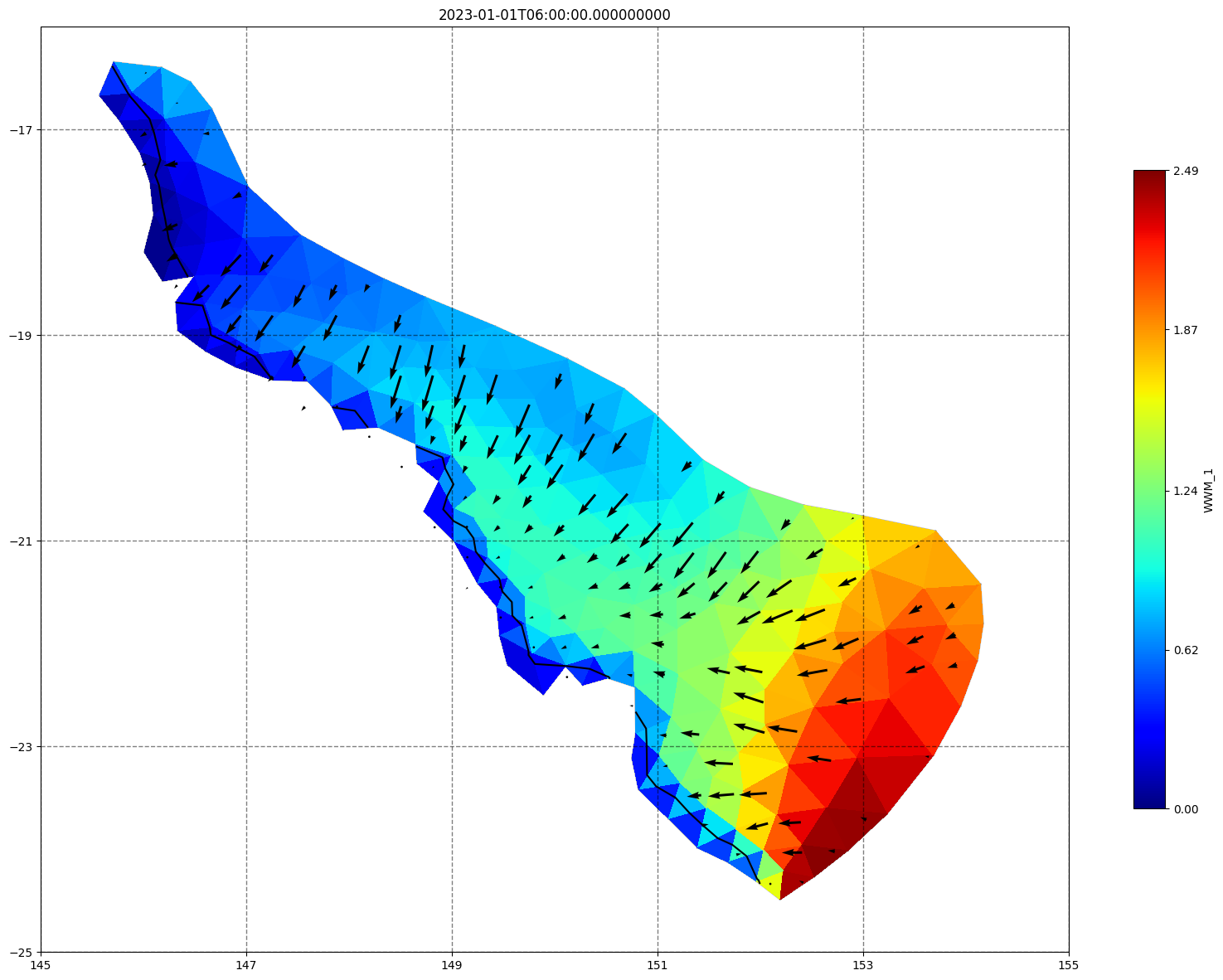
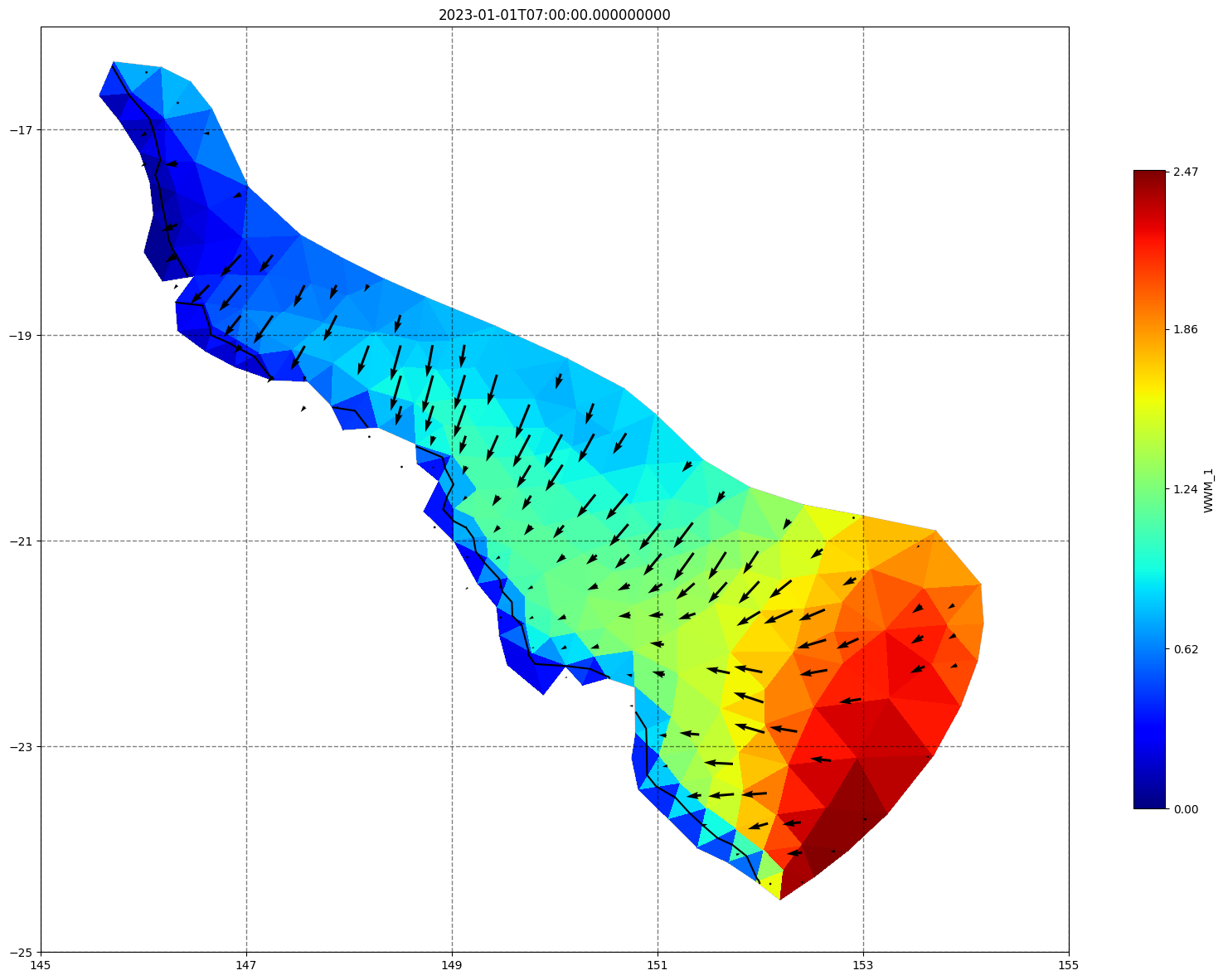
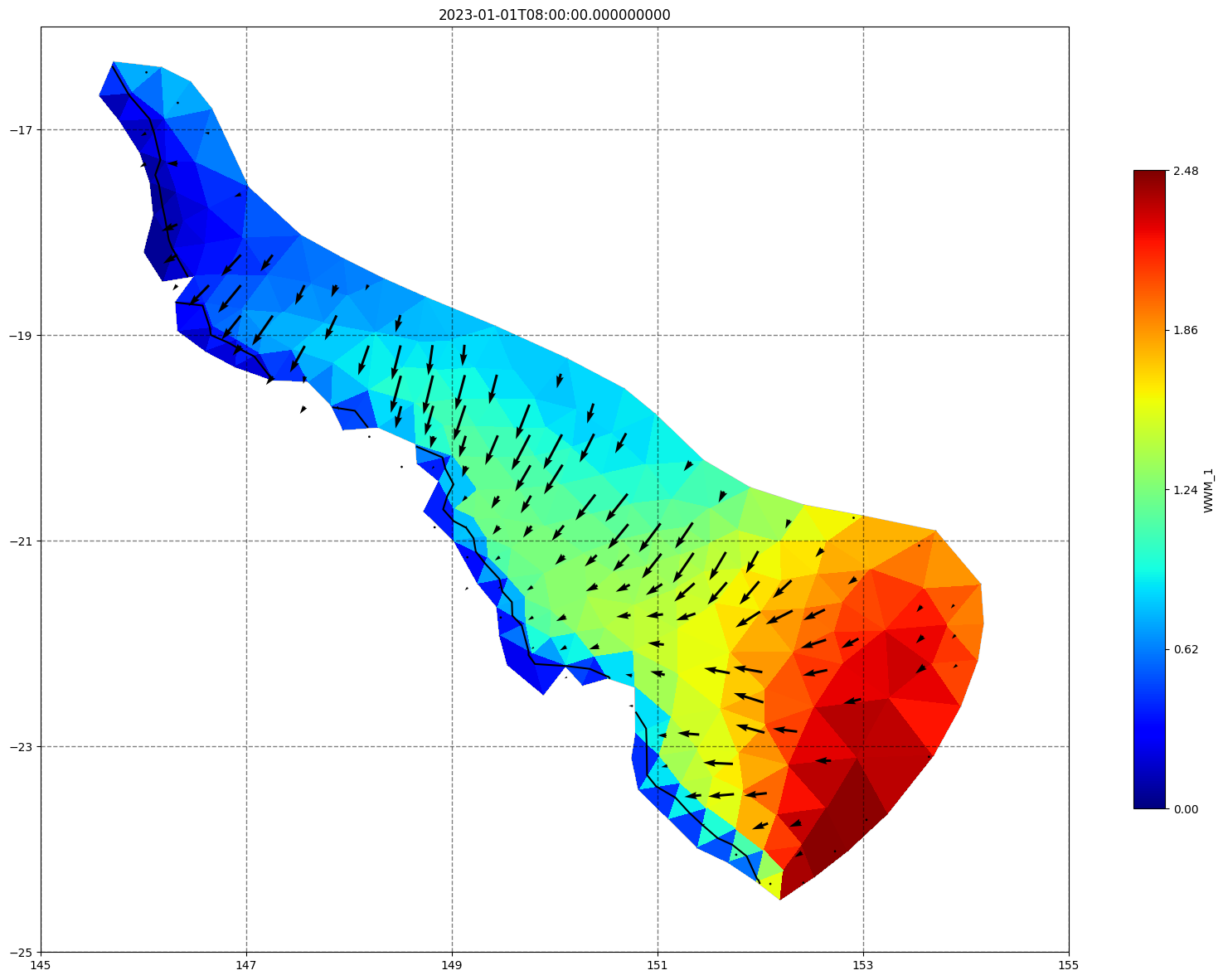
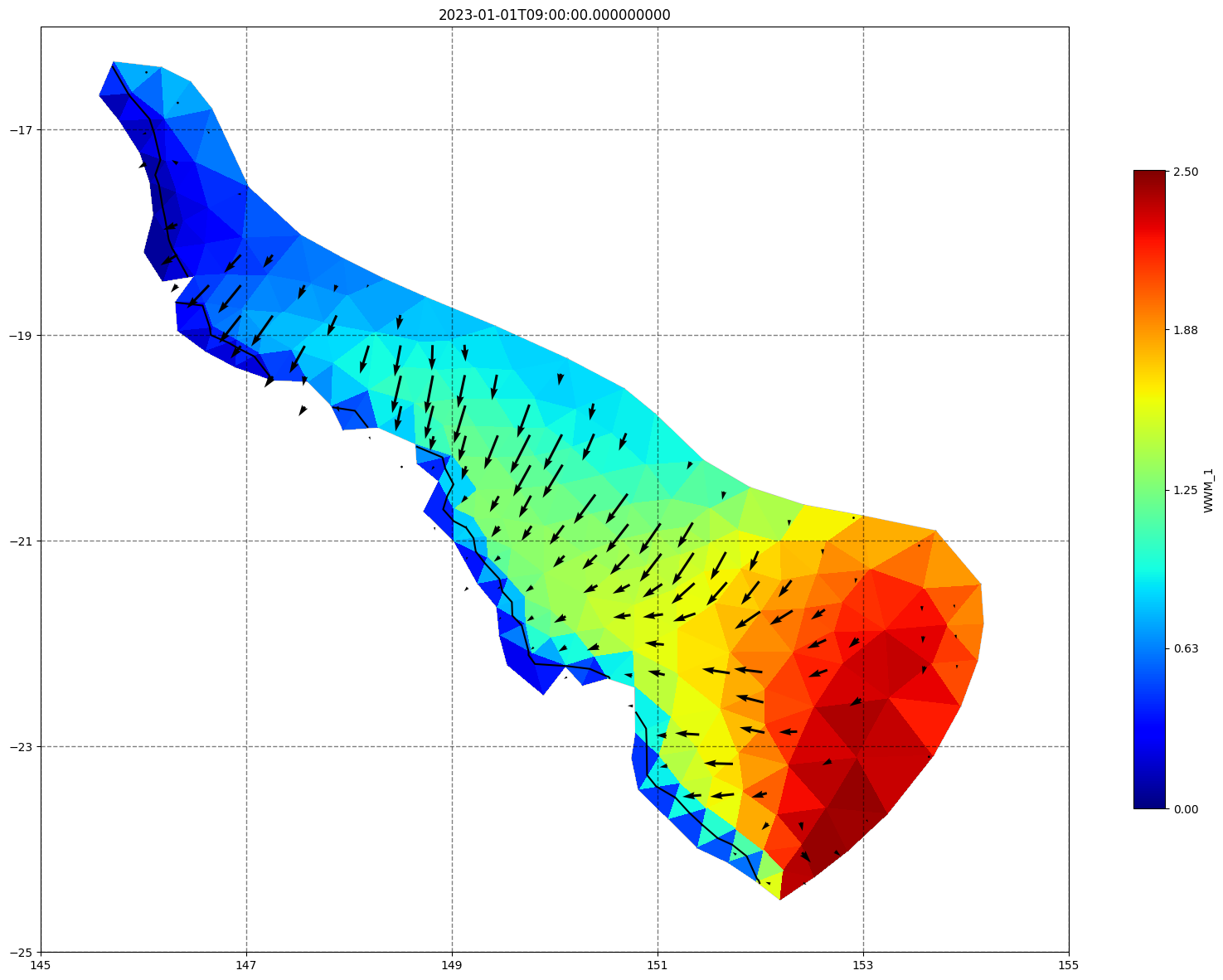
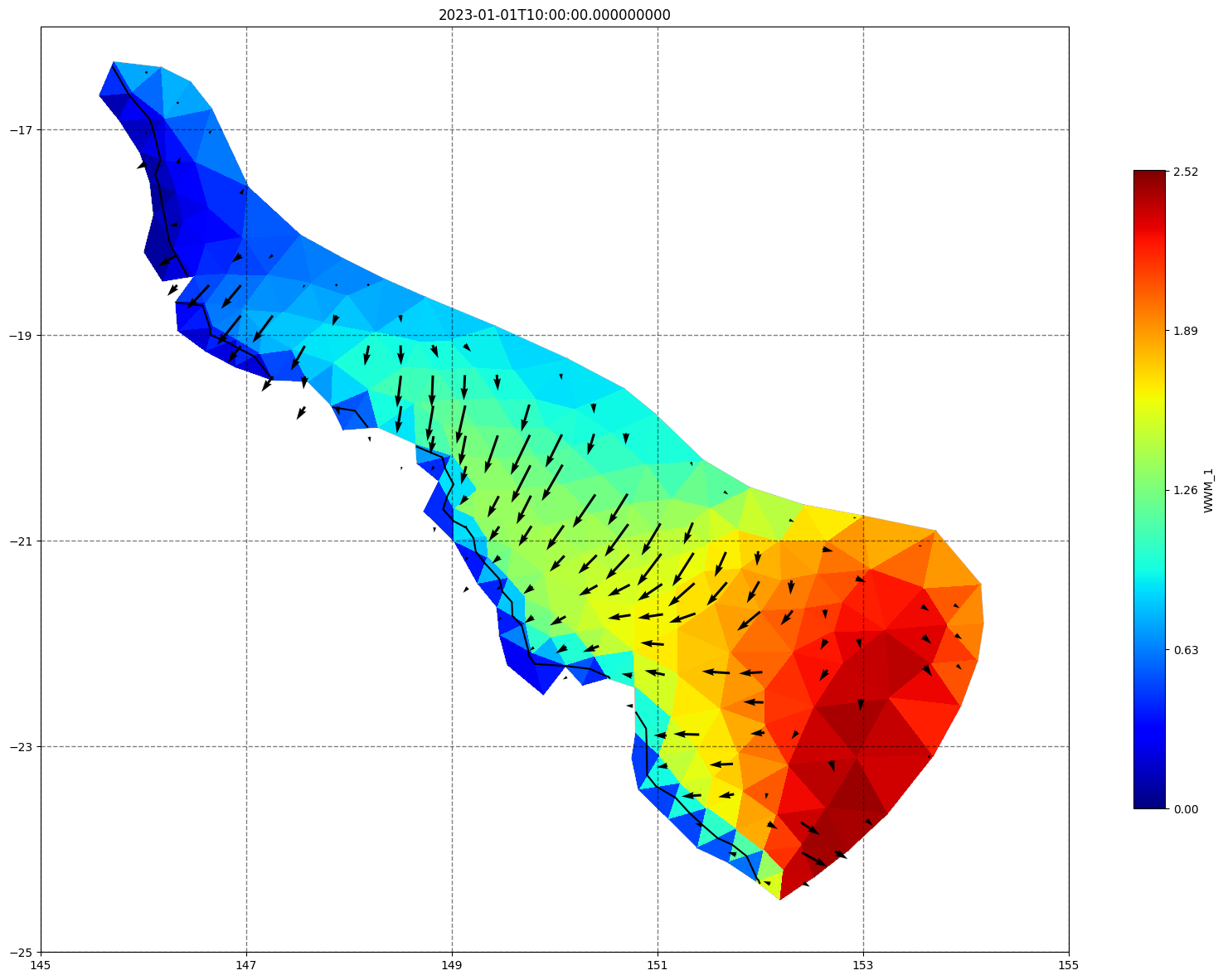
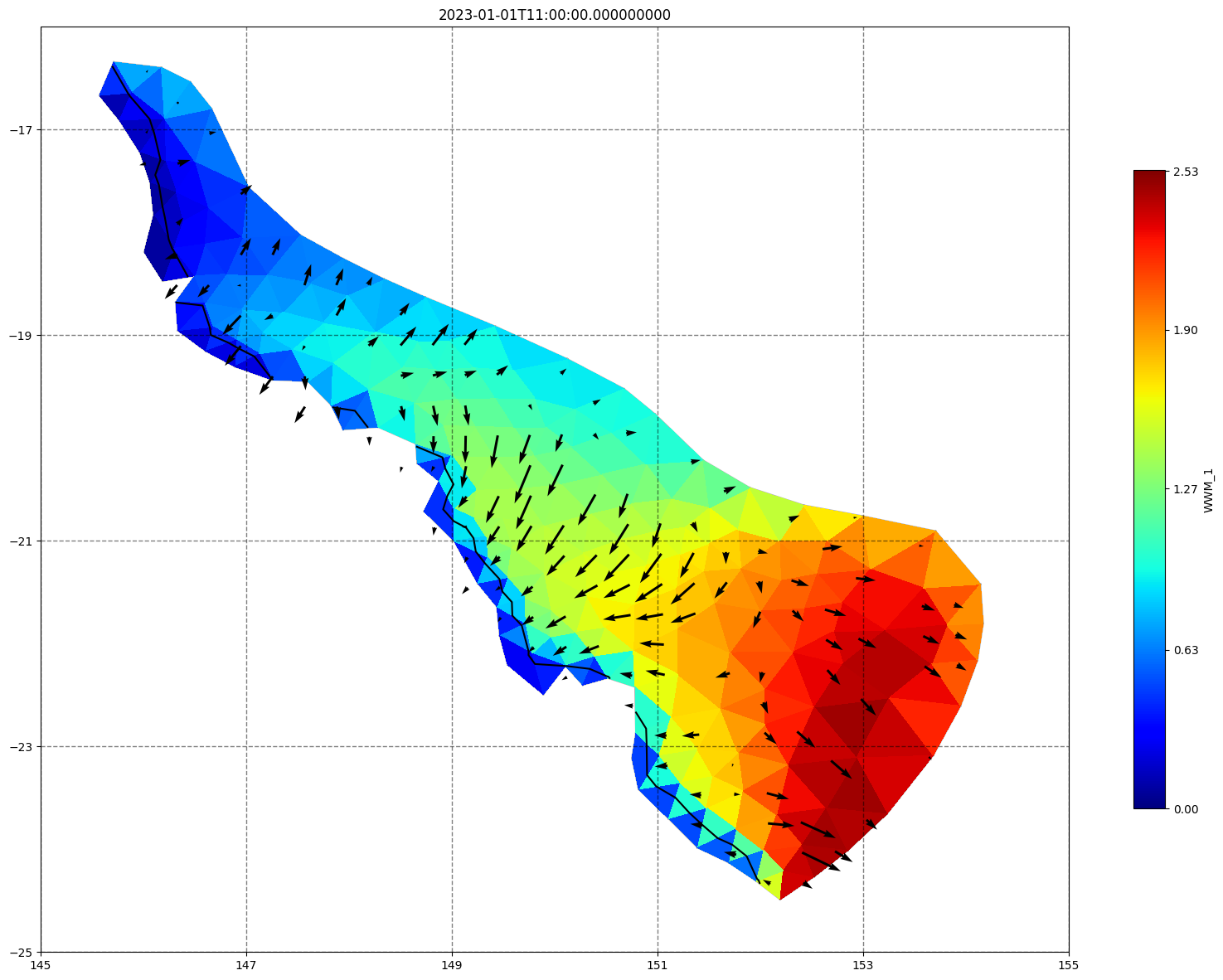
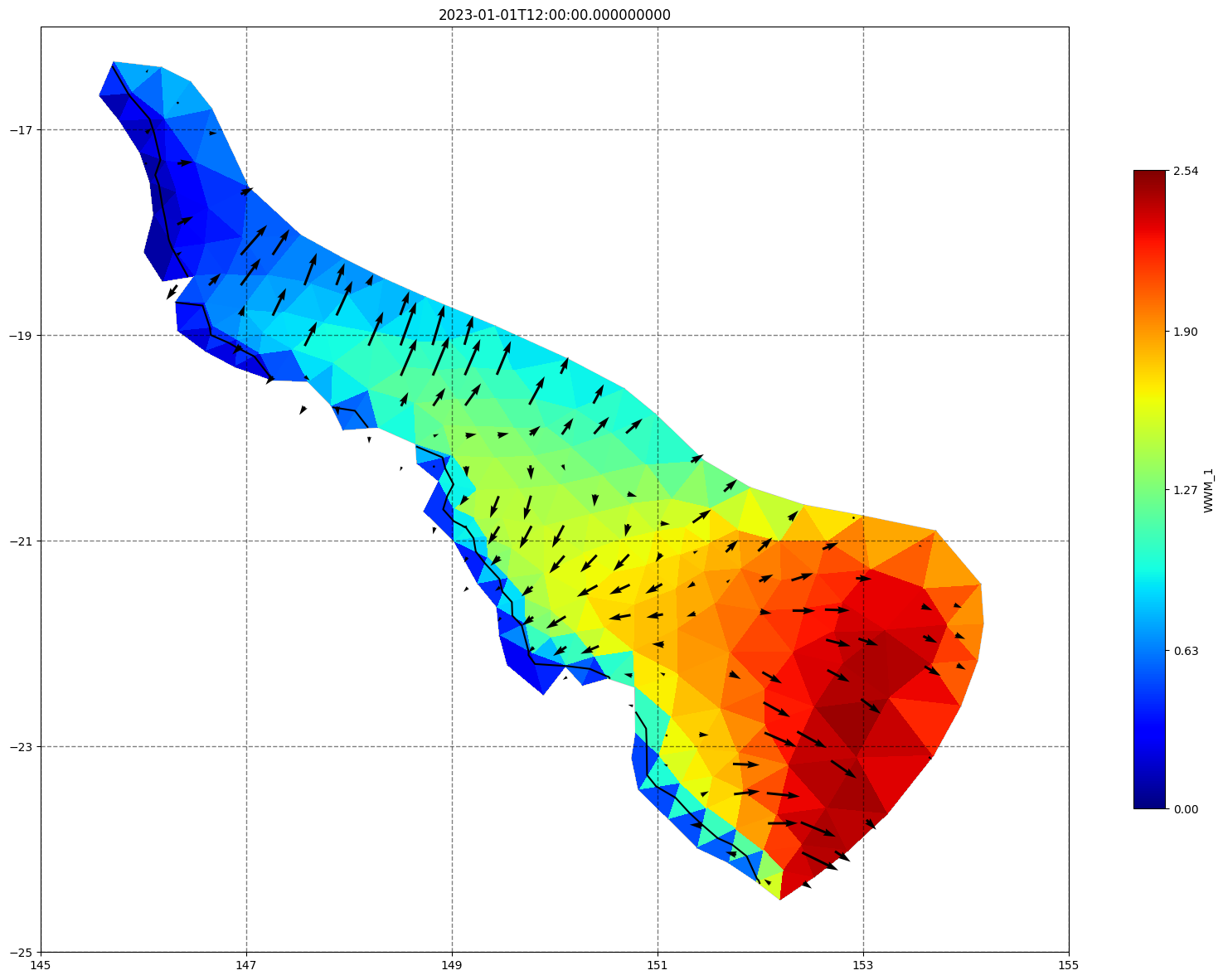
[35]:
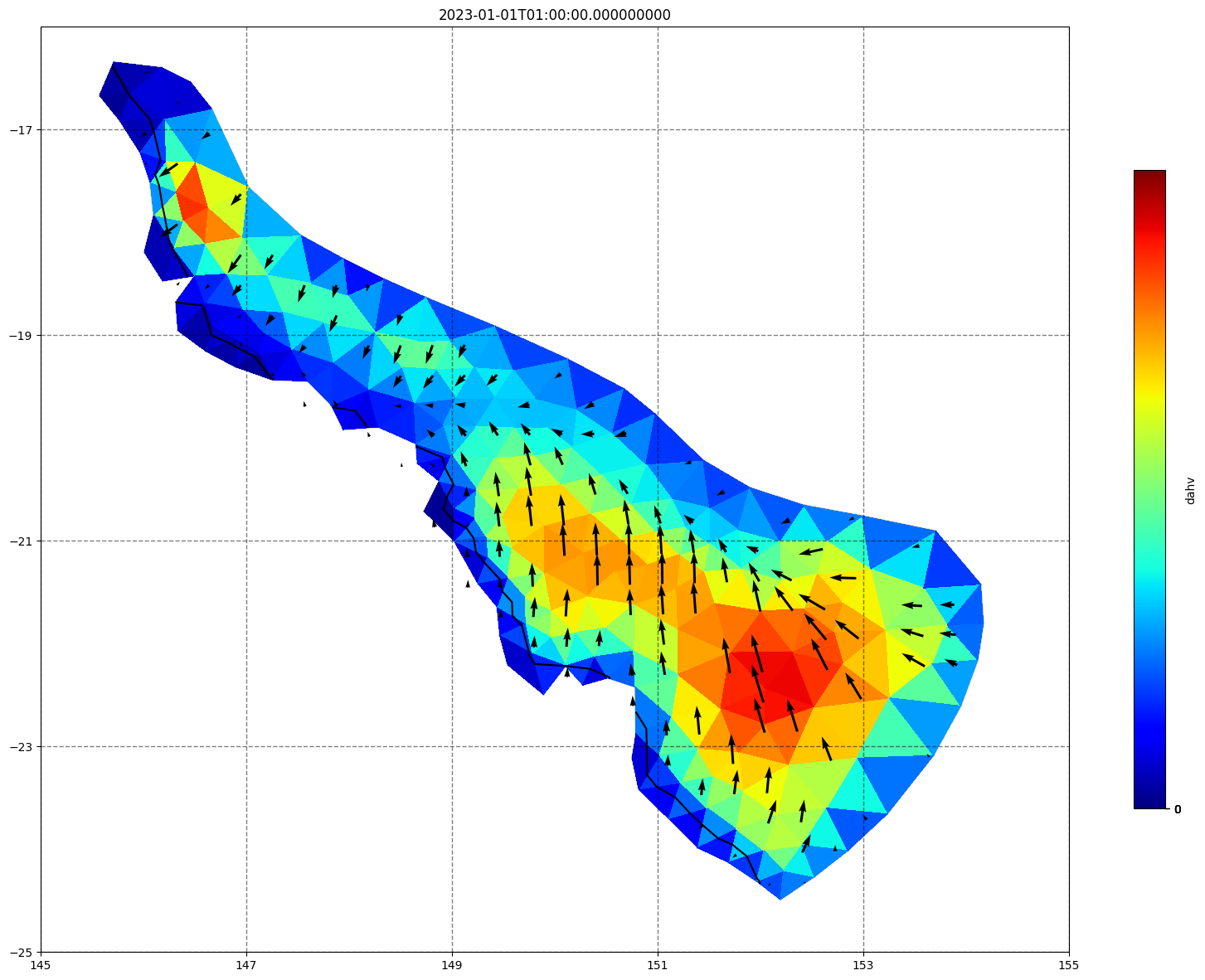
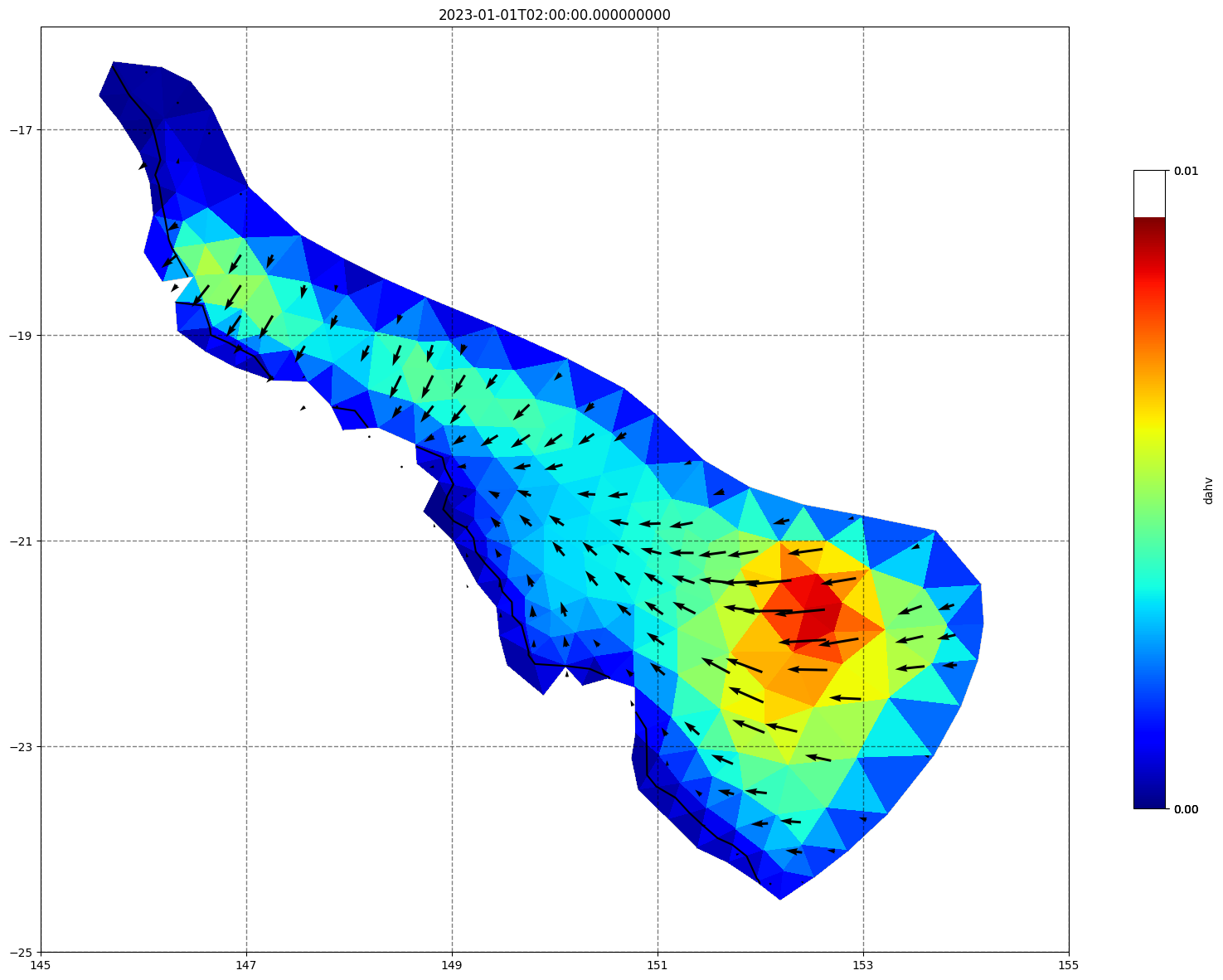
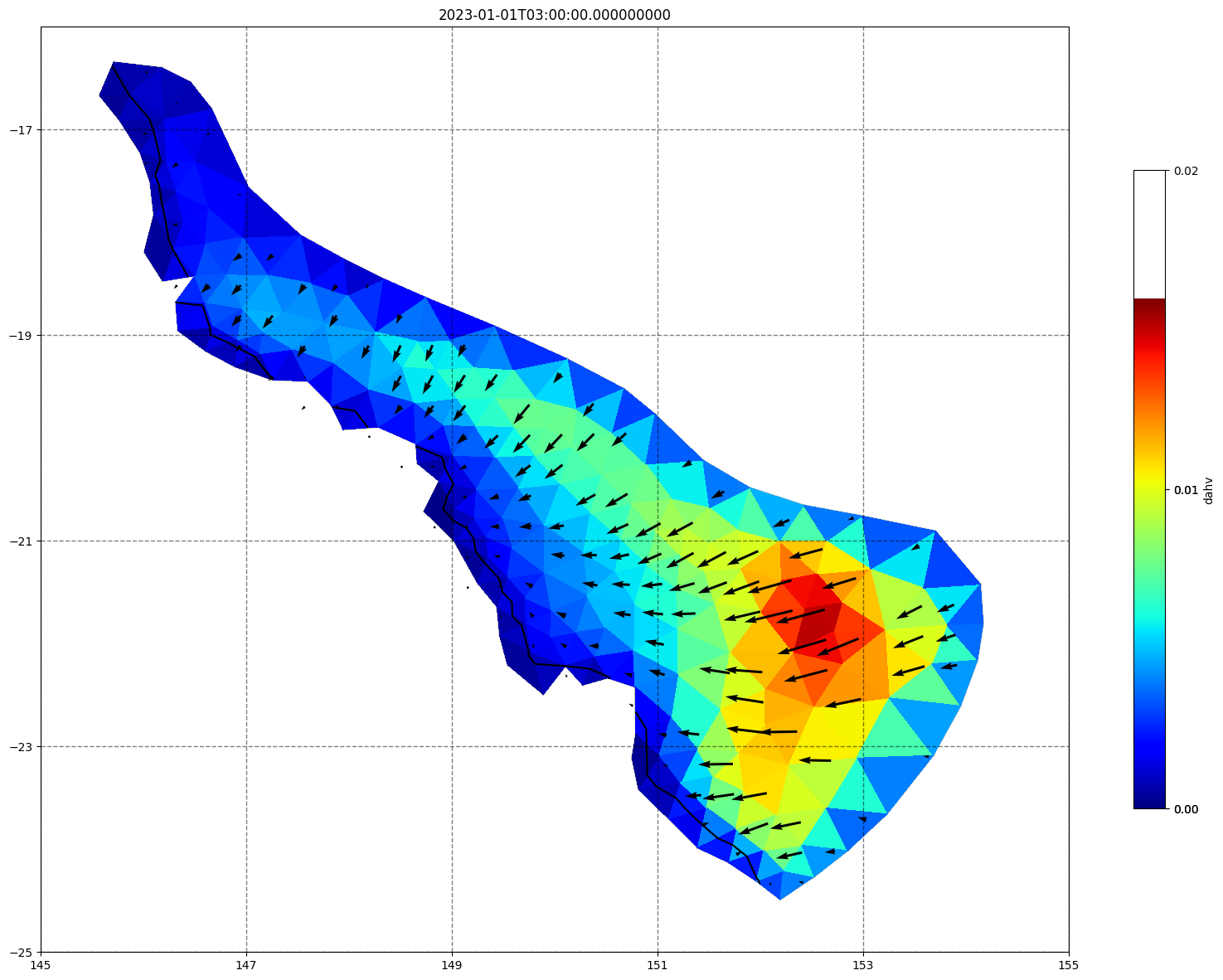
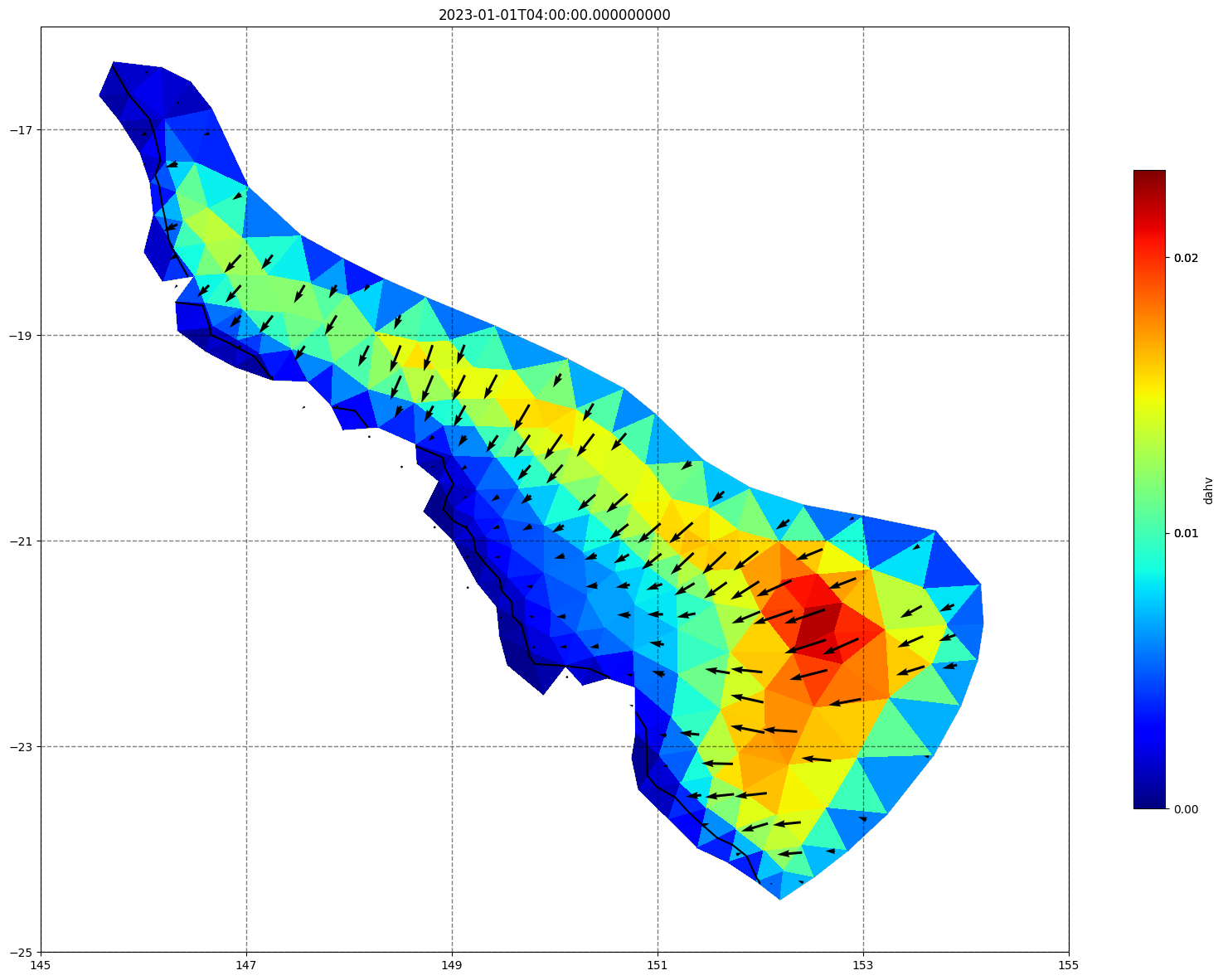
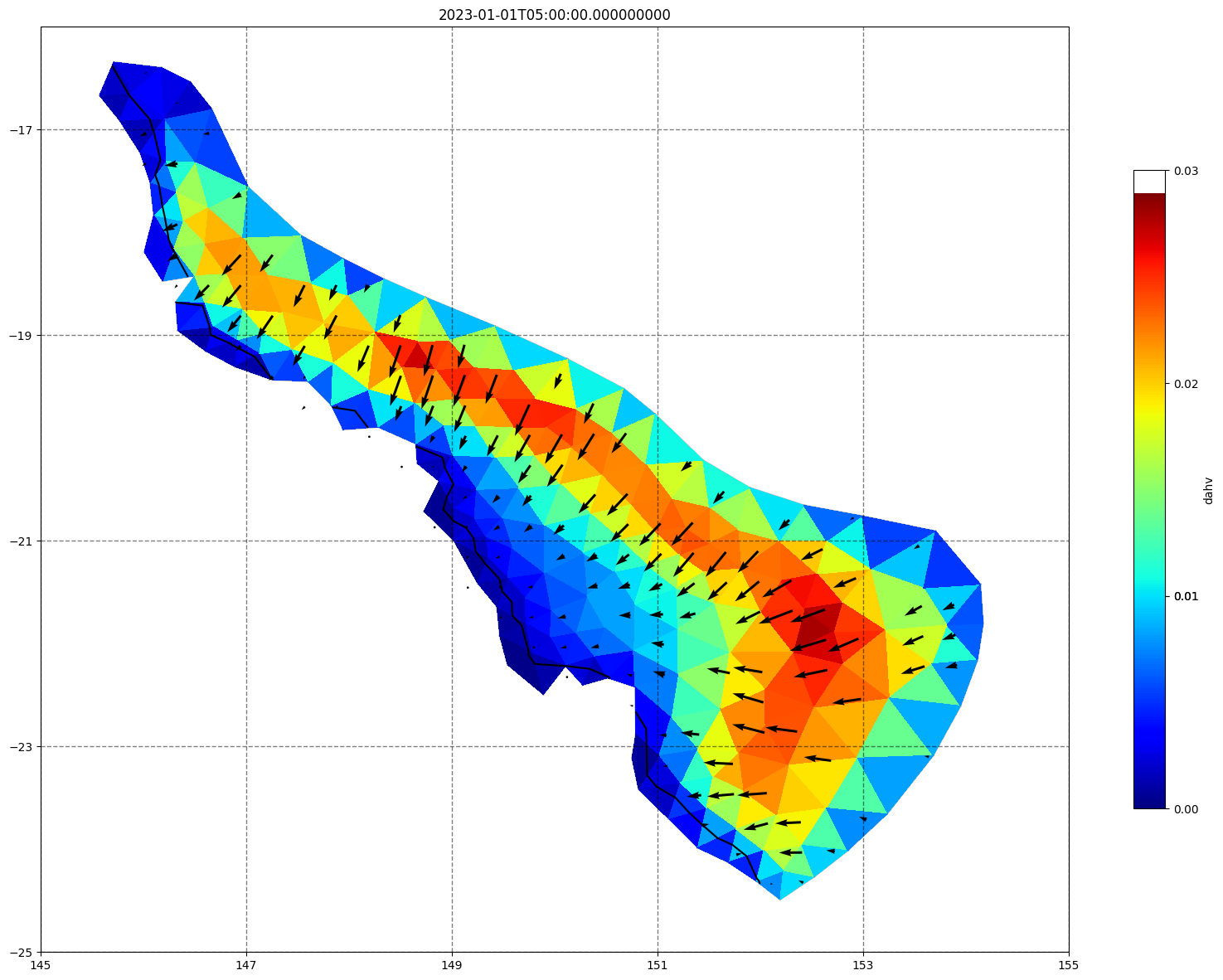
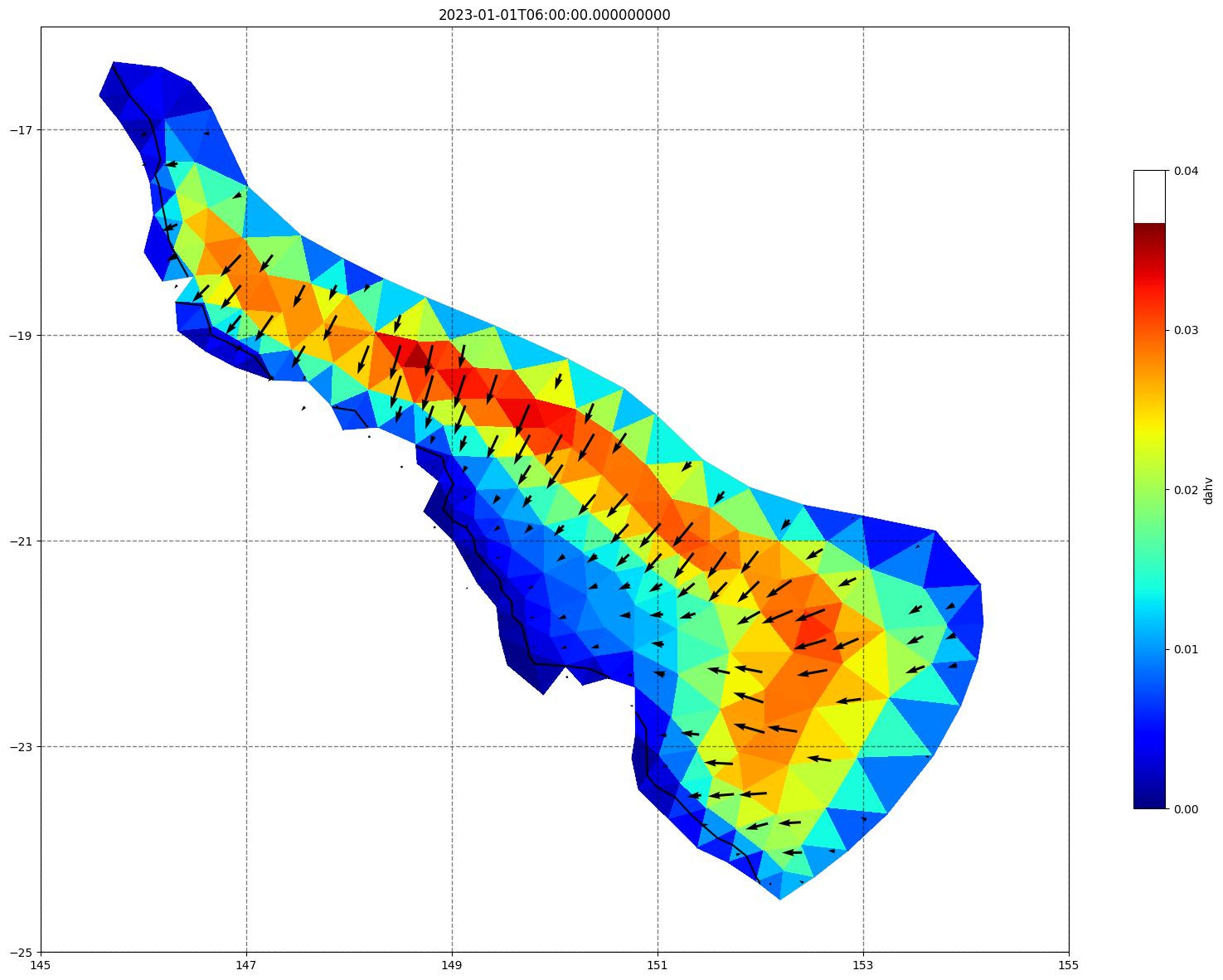
## plot gridded fields - Hs
for ix, time in enumerate(schout.time.values):
fig, ax=schism_plot(schout, meshtri,'dahv', bbox=[145,-25,155,-16], project=True, plotmesh=True, mask=False, time=time,
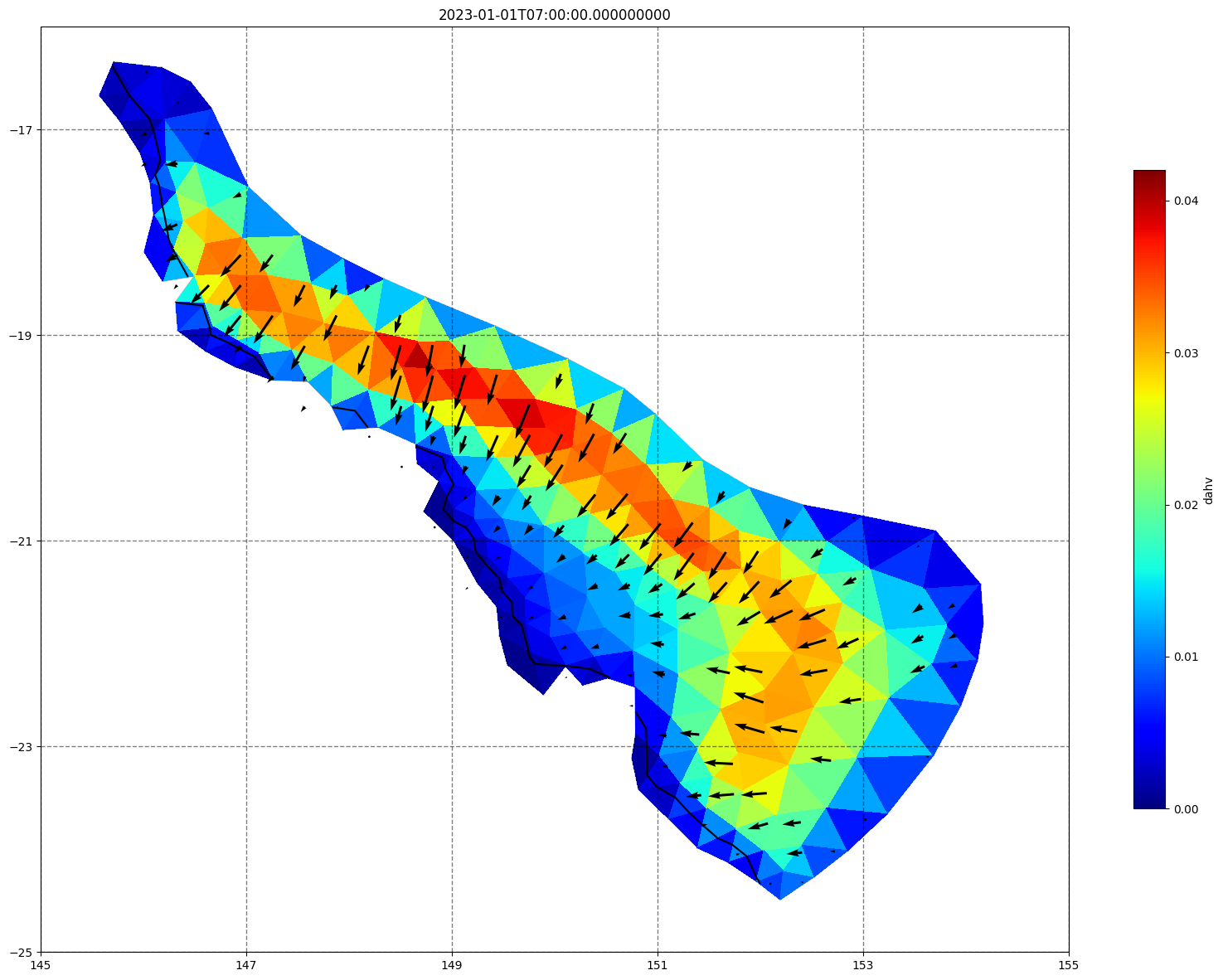
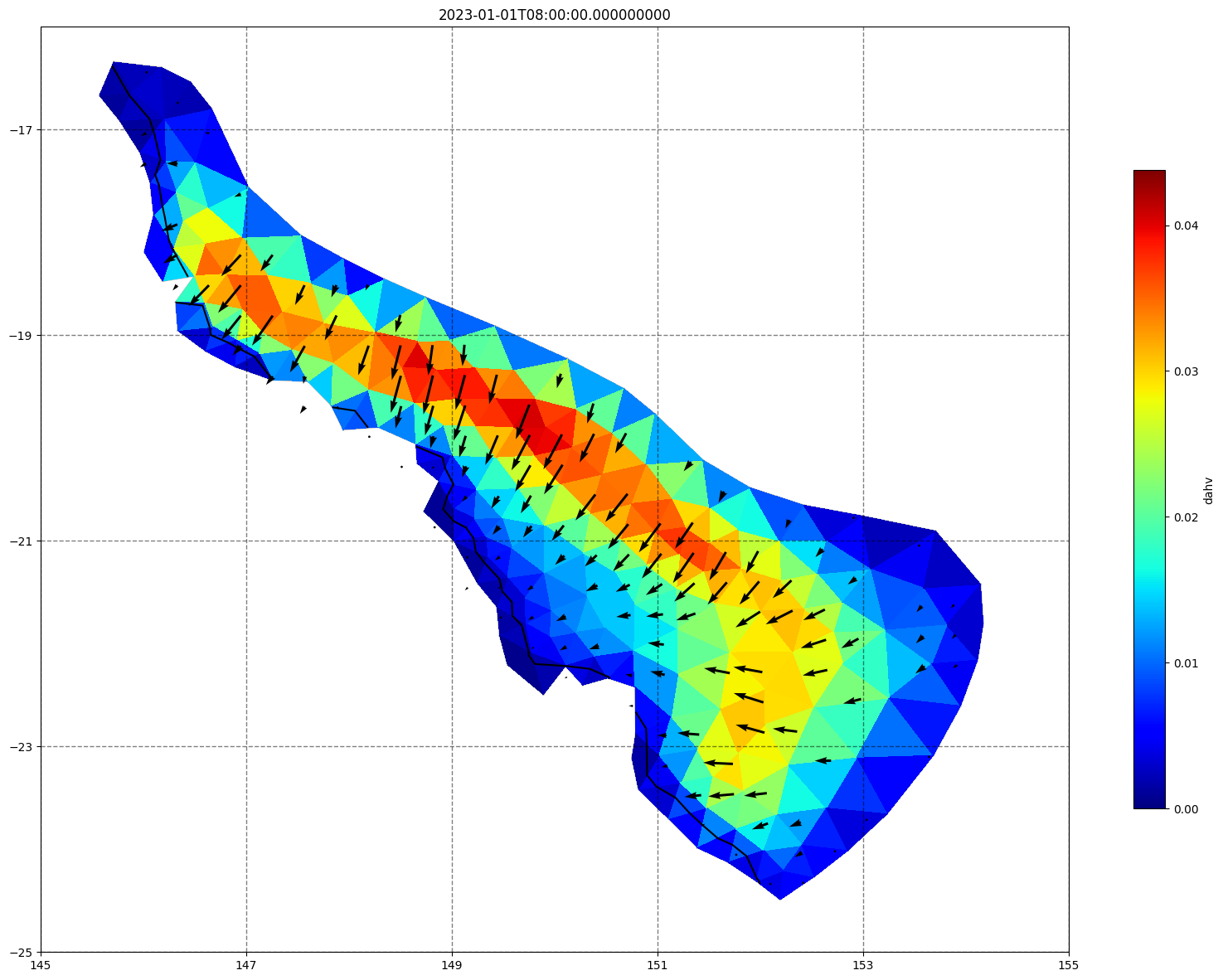
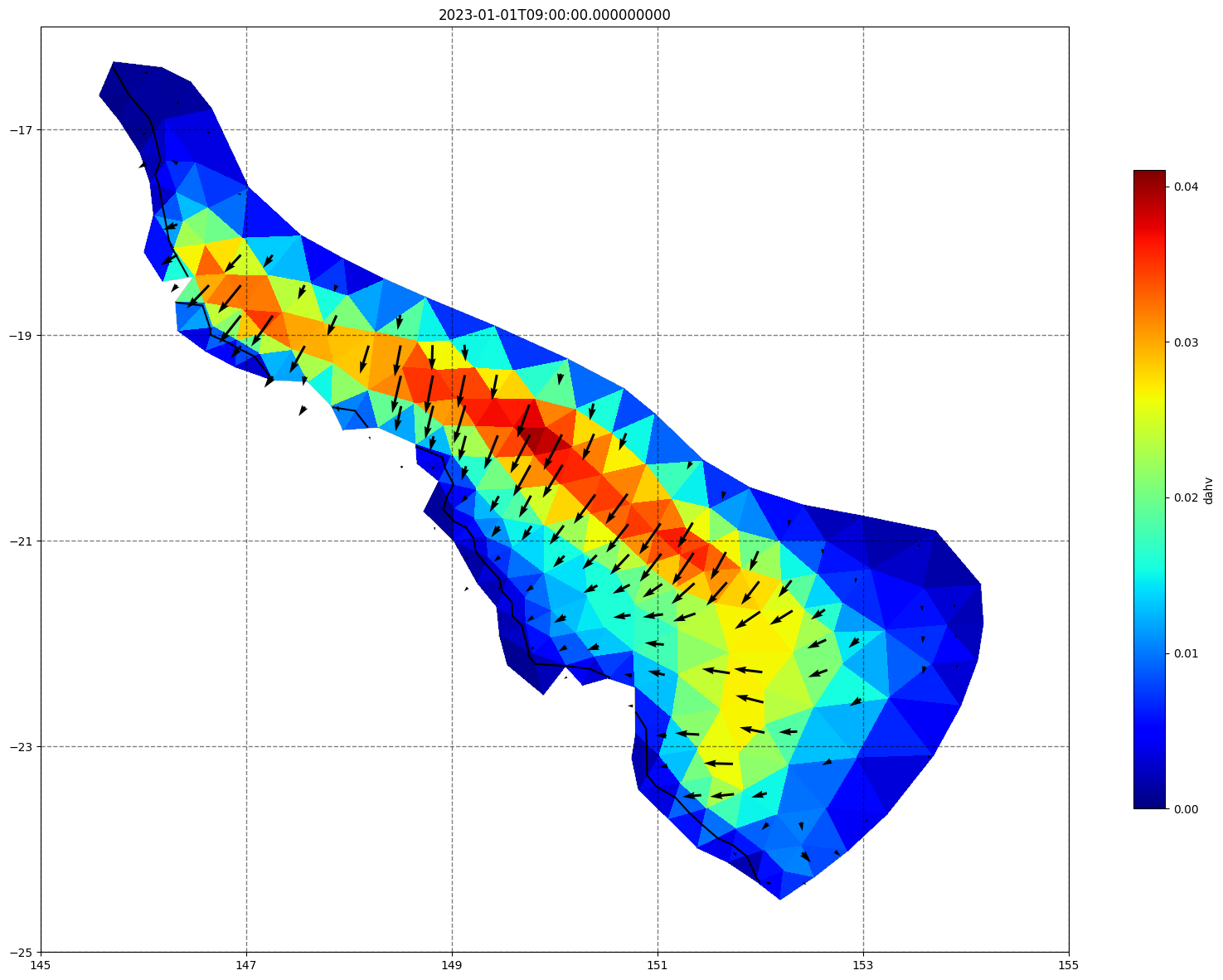
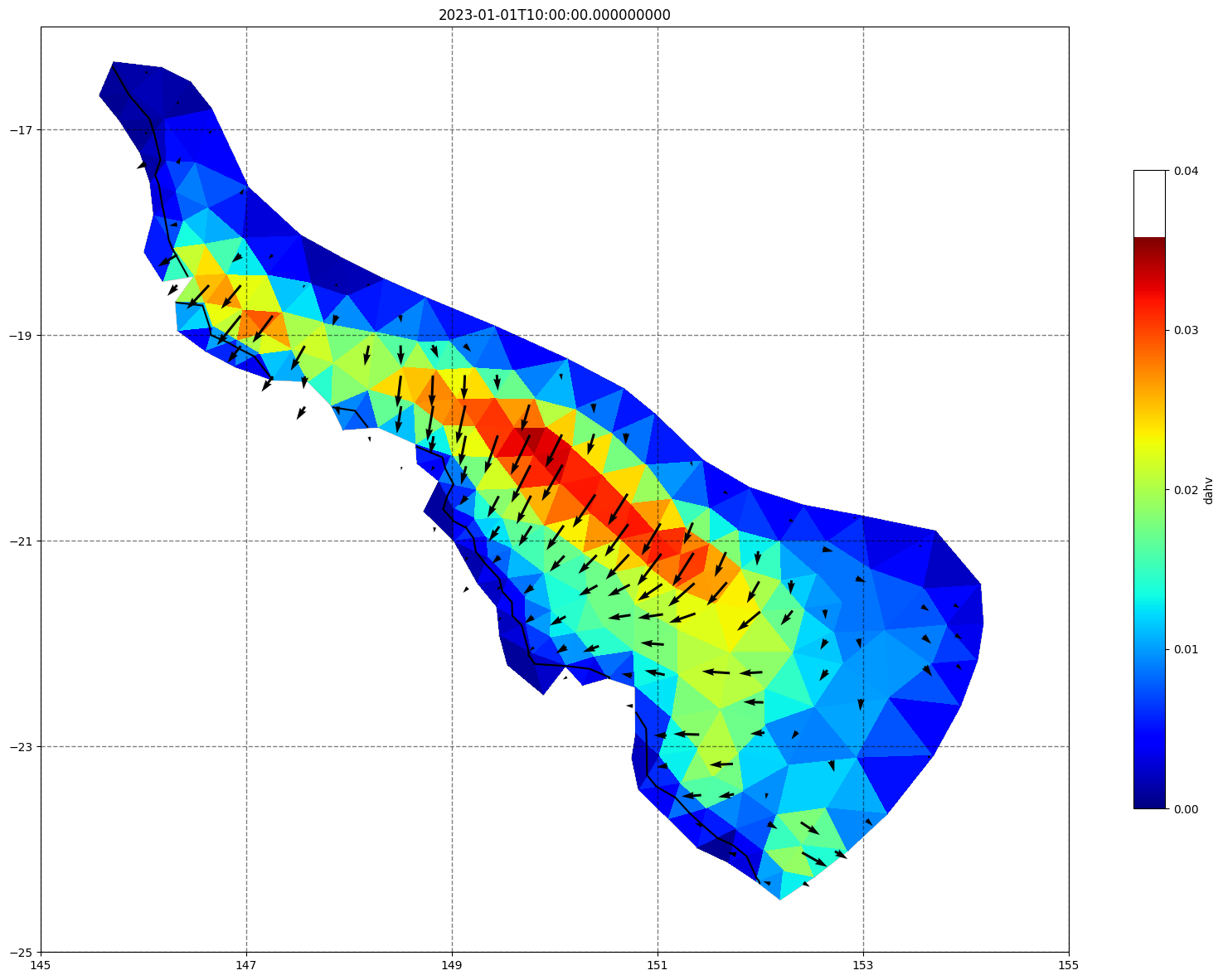
vectors=True, contours=[0])
ax.set_title(time)
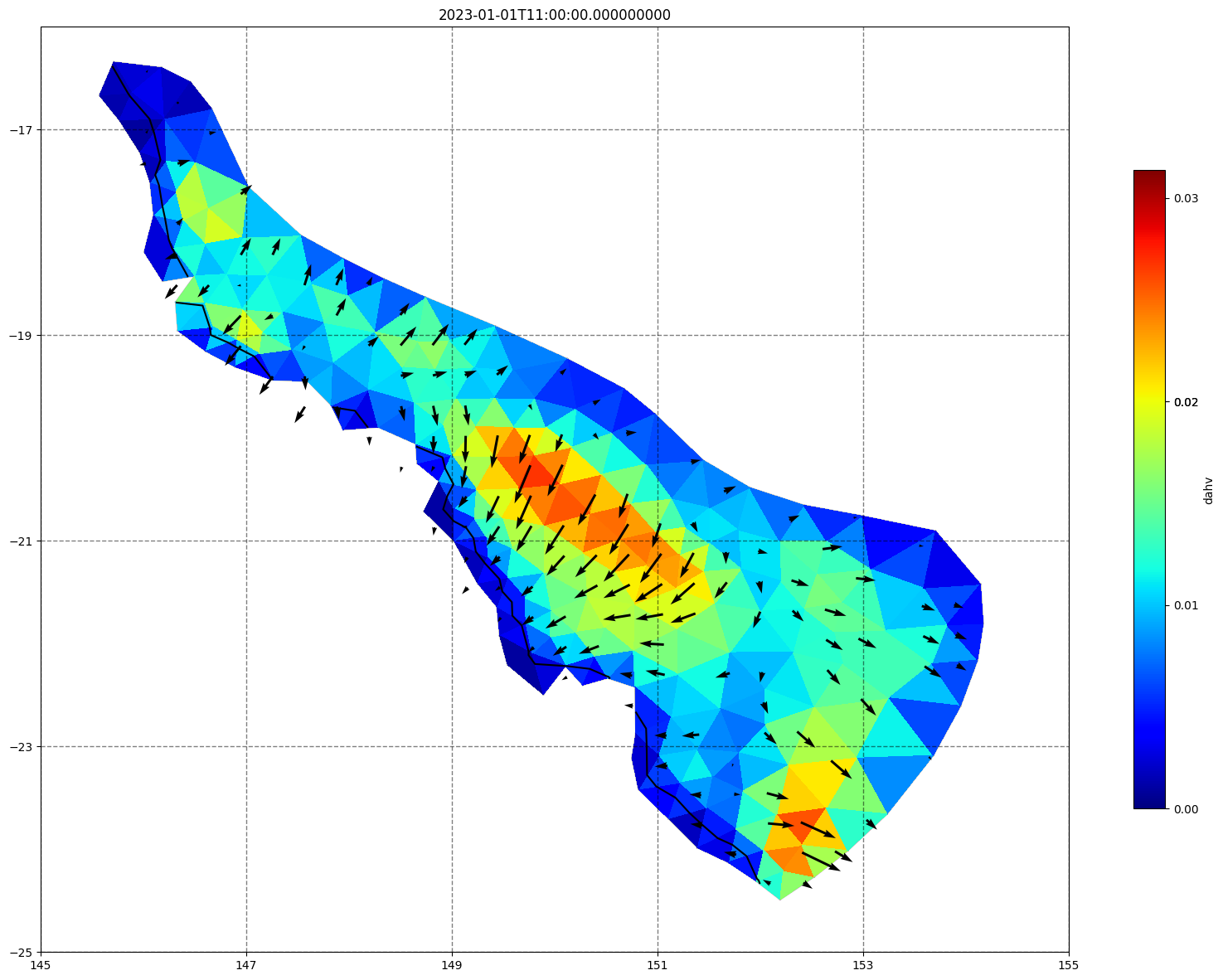
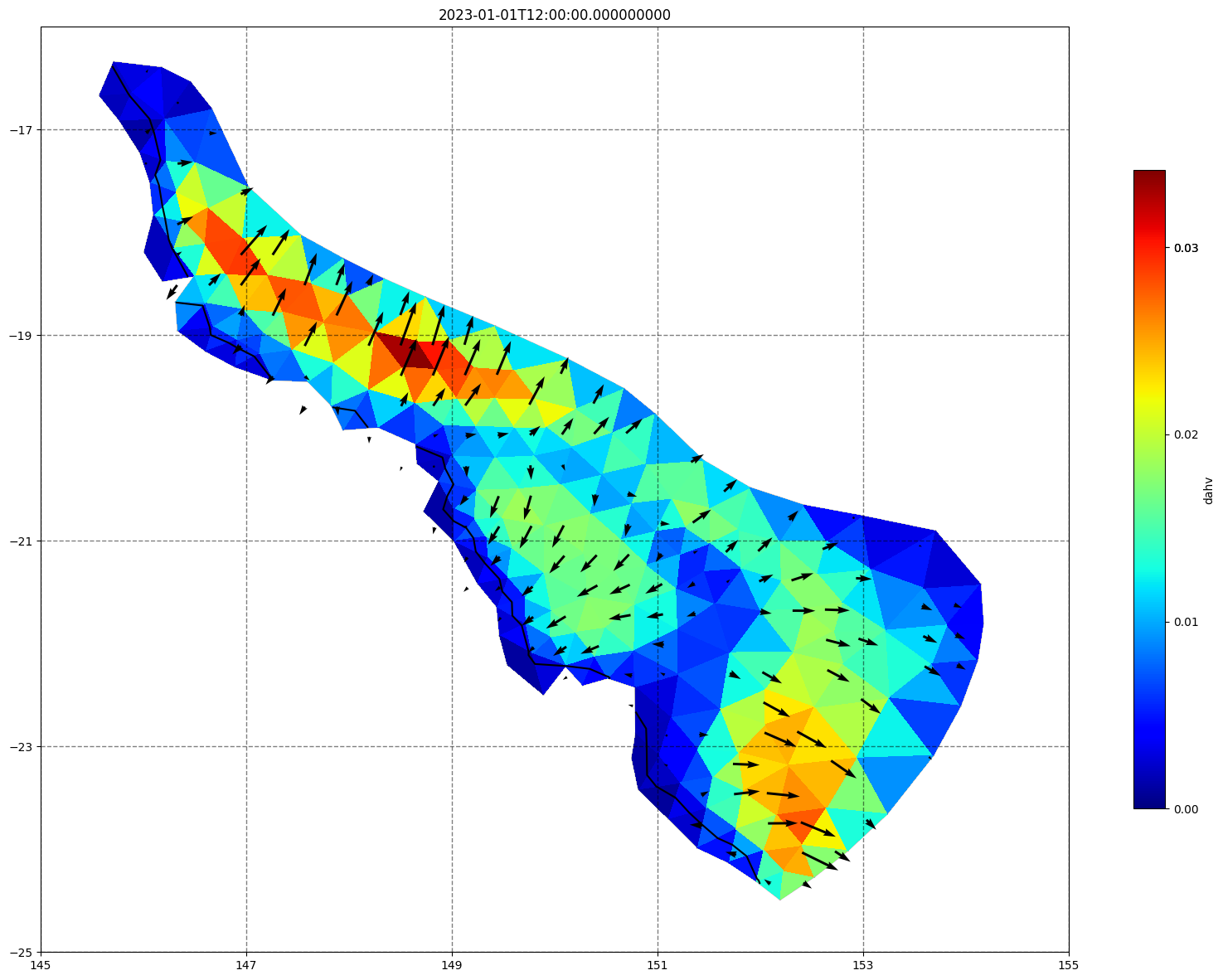
[ ]:
[36]:
# The full model can be dumped to a configuration file.
import yaml
# dump full model to yaml
with open('model.yaml', 'w') as f:
yaml.dump(run.model_dump(), f)
[37]:
!cat model.yaml
config:
CGPP: F
CPP: F
DM: T
DPEAK: T
DSPR: T
HS: T
KPP: F
LPP: F
ORBITAL: F
PEAKD: T
PEAKDSPR: T
TM01: T
TM02: F
TPP: T
TPPD: T
UBOT: F
WNPP: F
ac: T
brcrYN: ''
checkout: main
currYN: '!'
data:
atmos:
air_1:
buffer: 2.0
coords:
s: site
t: time
x: longitude
y: latitude
z: depth
crop_data: true
data_type: sflux_air
fail_if_missing: true
filter:
crop:
latitude:
start: -26.4884499831
stop: -14.343982374700001
longitude:
start: 143.572275673
stop: 156.1662613999
time:
start: &id001 2023-01-01 00:00:00
stop: 2023-01-01 13:00:00
derived: {}
rename: {}
sort:
coords:
- latitude
subset: {}
timenorm: {}
id: air_1
link: false
max_window_hours: 120.0
model_type: data_grid
prmsl_name: msl
relative_weight: 1.0
source:
kwargs: {}
model_type: file
uri: !!python/object/apply:pathlib.PosixPath
- ..
- ..
- tests
- schism
- test_data
- era5.nc
spfh_name: null
stmp_name: null
time_buffer:
- 0
- 1
uwind_name: u10
variables:
- u10
- v10
- msl
vwind_name: v10
air_2: null
data_type: sflux
prc_1: null
prc_2: null
rad_1: null
rad_2: null
data_type: schism
ocean:
SAL_3D: null
TEM_3D: null
data_type: ocean
elev2D:
buffer: 0.0
coords:
s: site
t: time
x: xlon
y: ylat
z: depth
crop_data: true
data_type: boundary
filter:
crop:
time:
start: *id001
stop: 2023-01-02 12:00:00
derived: {}
rename: {}
sort: {}
subset: {}
timenorm: {}
id: elev2D
interpolate_missing_coastal: true
link: false
model_type: data_boundary
sel_method: interp
sel_method_kwargs: {}
source:
kwargs: {}
model_type: file
uri: !!python/object/apply:pathlib.PosixPath
- ..
- ..
- tests
- schism
- test_data
- hycom.nc
spacing: null
time_buffer:
- 0
- 1
variable: surf_el
variables:
- surf_el
uv3D: null
tides:
add_earth_tidal: true
constituents:
- M2
- S2
- N2
cutoff_depth: 50.0
data_type: tide
database: tpxo
ethconst: []
flags:
- - 5
- 3
- 0
- 0
relax: []
sobc:
- 1
sthconst: []
tidal_data:
data_type: tidal_dataset
elevations: !!python/object/apply:pathlib.PosixPath
- ..
- ..
- tests
- schism
- test_data
- tpxo9-neaus
- h_m2s2n2.nc
velocities: !!python/object/apply:pathlib.PosixPath
- ..
- ..
- tests
- schism
- test_data
- tpxo9-neaus
- u_m2s2n2.nc
tobc:
- 1
tthconst: []
vthconst: []
wave:
buffer: 2.0
coords:
s: site
t: time
x: lon
y: lat
z: depth
crop_data: true
data_type: wave
filter:
crop:
time:
start: *id001
stop: 2023-01-01 18:00:00
derived: {}
rename: {}
sort: {}
subset: {}
timenorm: {}
grid_type: boundary_wave_station
id: wavedata
link: false
model_type: data_boundary
sel_method: nearest
sel_method_kwargs:
unique: true
source:
catalog_uri: !!python/object/apply:pathlib.PosixPath
- ..
- ..
- tests
- schism
- ..
- data
- catalog.yaml
catalog_yaml: null
dataset_id: ausspec
kwargs: {}
model_type: intake
spacing: null
time_buffer:
- 0
- 1
variables:
- efth
- lon
- lat
definetc: -1
deltc: 360
deltc_out: 3600
dramp: 1.0
drampwafo: 1.0
drampwind: 1.0
dzb_decayYN: '!'
extrap: T
extrapYN: '!'
filewind: wind.dat
fricc: 0.067
grid:
albedo:
crs: epsg:4326
gr3_type: albedo
hgrid:
id: hgrid
link: false
model_type: data_blob
source: &id002 !!python/object/apply:pathlib.PosixPath
- ..
- ..
- tests
- schism
- test_data
- hgrid_20kmto60km_rompyschism_testing.gr3
value: 0.15
crs: epsg:4326
diffmax:
crs: epsg:4326
gr3_type: diffmax
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
value: 1.0
diffmin:
crs: epsg:4326
gr3_type: diffmin
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
value: 1.0e-06
drag: null
grid_type: schism
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
hgrid_WWM:
gridtype: hgrid_WWM
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
hgridll:
gridtype: hgridll
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
manning:
crs: epsg:4326
gr3_type: manning
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
value: 1.0
rough: null
vgrid:
vgrid:
model_type: LSC2_generator
watertype:
crs: epsg:4326
gr3_type: watertype
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
value: 1.0
windrot_geo2proj:
crs: epsg:4326
gr3_type: windrot_geo2proj
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
value: 0.0
wwmbnd:
bcflags: null
hgrid:
id: hgrid
link: false
model_type: data_blob
source: *id002
h1_bcc: 50.0
h2_bcc: 100.0
h_bcc1: 100.0
ibreak: 1
ibtrack_openbndYN: '!'
ic_elev: 0
icou_elfe_wwm: 1
ihfskip: 720
ihot: 0
inv_atm_bnd: 1
iout_sta: 0
iouts: 15
iwbl: 0
iwind_form: 1
iwindoffYN: '!'
limfak: 0.1
lindsprdeg: F
lsourceswam: F
lsp2d: T
mdc2: 36
melim: 1
mesbf: 1
mesin: 1
model_type: schismcsiro
msc2: 36
nadv: 1
nchi: -1
nouts: '''AWAC_in'',''AWAC_mid'',''AWAC_off'',''SPOT_1002'',''SPOT_1011'',''SPOT_1018'',''SPOT_1026'''
nstep_wwm: 3
outstyle: NC
param_iof_hydro1: 1
param_iof_hydro10: 0
param_iof_hydro11: 0
param_iof_hydro12: 0
param_iof_hydro13: 0
param_iof_hydro14: 1
param_iof_hydro15: 0
param_iof_hydro16: 1
param_iof_hydro17: 0
param_iof_hydro18: 0
param_iof_hydro19: 0
param_iof_hydro2: 0
param_iof_hydro20: 0
param_iof_hydro21: 0
param_iof_hydro22: 0
param_iof_hydro23: 0
param_iof_hydro24: 0
param_iof_hydro25: 0
param_iof_hydro26: 0
param_iof_hydro27: 0
param_iof_hydro28: 0
param_iof_hydro29: 0
param_iof_hydro3: 0
param_iof_hydro30: 0
param_iof_hydro4: 0
param_iof_hydro5: 0
param_iof_hydro6: 0
param_iof_hydro7: 0
param_iof_hydro8: 0
param_iof_hydro9: 0
param_nhot: 0
param_nhot_write: 22320.0
param_nhot_writeYN: '!'
param_nspool_sta: 30
project: WAXA
rlatitude: -29
sav_cdYN: '!'
sfea0: -29.0
slam0: 120.0
template: /home/tdurrant/source/rompy/rompy-base/rompy/templates/schismcsiro
thetai: 0.8
time_step: 120.0
utc_start: 0
walvYN: '!'
wbdm: 90
windYN: '!'
wwm1: 1
wwm10: 0
wwm11: 0
wwm12: 0
wwm13: 0
wwm14: 0
wwm15: 0
wwm16: 1
wwm17: 1
wwm18: 1
wwm19: 0
wwm2: 1
wwm20: 0
wwm21: 0
wwm22: 0
wwm23: 0
wwm24: 0
wwm25: 0
wwm26: 0
wwm27: 0
wwm28: 0
wwm29: 0
wwm3: 0
wwm30: 0
wwm31: 0
wwm31YN: '!'
wwm32: 0
wwm32YN: '!'
wwm33: 0
wwm33YN: '!'
wwm34: 0
wwm34YN: '!'
wwm35: 0
wwm35YN: '!'
wwm36: 0
wwm36YN: '!'
wwm37: 0
wwm37YN: '!'
wwm4: 0
wwm5: 0
wwm6: 0
wwm7: 0
wwm8: 1
wwm9: 1
wwminput_LHOTF: F
wwminput_LHOTR: F
wwminput_LINID: T
wwminput_history_DELTC: 3600
wwminput_history_DEP: F
wwminput_history_OUTSTYLE: NC
wwminput_history_STOKESBAROX: T
wwminput_history_STOKESBAROY: T
wwminput_history_STOKESSURFX: T
wwminput_history_STOKESSURFY: T
wwminput_history_TAUHF: T
wwminput_history_TAUTOT: T
wwminput_history_TAUW: F
wwminput_hotfile_DELTC: 0
wwminput_station_DELTC: 3600
wwminput_station_DEP: T
wwminput_station_OUTSTYLE: 'NO'
wwminput_station_STOKESBAROX: T
wwminput_station_STOKESBAROY: T
wwminput_station_STOKESSURFX: T
wwminput_station_STOKESSURFY: T
wwminput_station_TAUHF: T
wwminput_station_TAUTOT: T
wwminput_station_TAUW: T
xouts: 115.6208687,115.5941886,115.58077,115.5942931,115.5830497,115.5807825,115.5960683
youts: -32.611605,-32.611605,-32.613682,-32.6253914,-32.6135870,-32.6294226,-32.6096741
output_dir: !!python/object/apply:pathlib.PosixPath
- schism_procedural
period:
duration: !!python/object/apply:datetime.timedelta
- 0
- 43200
- 0
end: 2023-01-01 12:00:00
include_end: true
interval: !!python/object/apply:datetime.timedelta
- 0
- 3600
- 0
start: *id001
run_id: test_schism
Running from configuration files.#
The full model dump above looks complex due to the fact that the full model state, including all default value, is written to the model.yaml file. The same model configuration can be achived in a much simpler file by simply specifying non default values. For example, the entire configuration above is specified in the demo.yaml file shown below
[38]:
!cat demo.yaml
output_dir: schism_declaritive
period:
start: 20230101T00
end: 20230101T12
interval: 3600
run_id: test_schismcsiro
config:
model_type: schismcsiro
mesbf: 1
fricc: 0.067
param_iof_hydro1: 1 # elevation
param_iof_hydro2: 1 # mslp
param_iof_hydro14: 1 # wind speed
param_iof_hydro16: 1 # surface velocities
wwm18: 1 # peak wave direction
wwm1: 1 # significant wave height
wwm9: 1 # peak period
grid:
grid_type: schism
hgrid:
id: hgrid
model_type: data_blob
#source: ../../tests/schism/test_data/hgrid.gr3
source: ../../tests/schism/test_data/hgrid_20kmto60km_rompyschism_testing.gr3
manning: 1
data:
data_type: schism
atmos:
air_1:
data_type: sflux_air
source:
model_type: file
uri: "../../tests/schism/test_data/era5.nc"
uwind_name: u10
vwind_name: v10
prmsl_name: msl
filter:
sort: {coords: [latitude]}
buffer: 5
ocean:
elev2D:
buffer: 0.0
coords:
t: time
x: xlon
y: ylat
z: depth
source:
uri: ../../tests/schism/test_data/hycom.nc
model_type: file
variable: surf_el
tides:
constituents:
- M2
- S2
- N2
cutoff_depth: 50.0
flags:
- [5, 3, 0, 0]
tidal_data:
data_type: tidal_dataset
elevations: ../../tests/schism/test_data/tpxo9-neaus/h_m2s2n2.nc
velocities: ../../tests/schism/test_data/tpxo9-neaus/u_m2s2n2.nc
wave:
buffer: 0.0
coords:
t: time
x: lon
y: lat
z: depth
id: wavedata
source:
catalog_uri: ../../tests/data/catalog.yaml
dataset_id: ausspec
model_type: intake
[39]:
# This can be loaded and used to instatiate the model object and run as above, e.g
import yaml
demo_config = yaml.load(open('demo.yaml', 'r'), Loader=yaml.FullLoader)
run = ModelRun(**demo_config)
# Remove old run if it exists
if run.output_dir.exists():
rmtree(run.output_dir)
run()
WARNING:rompy.schism.grid:manning is being set to a constant value, this is not recommended. For best results, please supply friction gr3 files with spatially varying values. Further options are under development.
INFO:rompy.model:
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Model settings:
INFO:rompy.model:
run_id: test_schismcsiro
period:
Start: 2023-01-01 00:00:00
End: 2023-01-01 12:00:00
Duration: 12:00:00
Interval: None
Include End: True
output_dir: schism_declaritive
config: <class 'rompy.schism.config.SchismCSIROConfig'>
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Generating model input files in schism_declaritive
INFO:rompy.schism.grid:Generated albedo with constant value of 0.15
INFO:rompy.schism.grid:Generated diffmin with constant value of 1e-06
INFO:rompy.schism.grid:Generated diffmax with constant value of 1.0
INFO:rompy.schism.grid:Generated watertype with constant value of 1.0
INFO:rompy.schism.grid:Generated windrot_geo2proj with constant value of 0.0
INFO:rompy.schism.grid:Generated manning with constant value of 1.0
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_declaritive/test_schismcsiro/hgrid.ll
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_declaritive/test_schismcsiro/hgrid_WWM.gr3
INFO:rompy.schism.data:Fetching air_1
INFO:rompy.schism.data:Fetching elev2D
INFO:rompy.schism.data:Fetching wavedata
INFO:rompy.schism.data:Generating tides
INFO:pyschism.forcing.bctides.bctides:Processing boundary 1:
INFO:pyschism.forcing.bctides.bctides:Elevation type: 5
WARNING:pyschism.forcing.bctides.bctides:Combination of 3 and 4, time history of elevation is read in from elev2D.th.nc!
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent M2.
INFO:pyschism.forcing.bctides.tpxo:h_file is ../../tests/schism/test_data/tpxo9-neaus/h_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent N2.
INFO:pyschism.forcing.bctides.bctides:Velocity type: 3
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent M2.
INFO:pyschism.forcing.bctides.tpxo:u_file is ../../tests/schism/test_data/tpxo9-neaus/u_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent N2.
INFO:pyschism.forcing.bctides.bctides:Temperature type: 0
WARNING:pyschism.forcing.bctides.bctides:Temperature is not sepcified, not input needed!
INFO:pyschism.forcing.bctides.bctides:Salinity type: 0
INFO:pyschism.forcing.bctides.bctides:Salinity is not sepcified, not input needed!
INFO:rompy.model:
INFO:rompy.model:Successfully generated project in schism_declaritive
INFO:rompy.model:-----------------------------------------------------
[39]:
'/home/tdurrant/source/rompy/rompy-base/notebooks/schism/schism_declaritive/test_schismcsiro'
[40]:
# Alternatively, this same config can be run directly using the rompy cli
!rm -fr schism_declaritive #remove previous run
!rompy schism demo.yaml
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/pyschism/forcing/hycom/gofs.py:8: UserWarning: The seawater library is deprecated! Please use gsw instead.
import seawater as sw
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/pydantic/_internal/_model_construction.py:64: UserWarning: `validate_rheol_steps` overrides an existing Pydantic `@model_validator` decorator
warnings.warn(f'`{k}` overrides an existing Pydantic `{existing.decorator_info.decorator_repr}` decorator')
WARNING:rompy.schism.grid:manning is being set to a constant value, this is not recommended. For best results, please supply friction gr3 files with spatially varying values. Further options are under development.
INFO:rompy.model:
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Model settings:
INFO:rompy.model:
run_id: test_schismcsiro
period:
Start: 2023-01-01 00:00:00
End: 2023-01-01 12:00:00
Duration: 12:00:00
Interval: None
Include End: True
output_dir: schism_declaritive
config: <class 'rompy.schism.config.SchismCSIROConfig'>
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Generating model input files in schism_declaritive
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/pydantic/main.py:426: UserWarning: Pydantic serializer warnings:
PydanticSerializationUnexpectedValue: Expected `BaseConfig` but got `SchismCSIROConfig` with value `SchismCSIROConfig(model_t...nput_history_DELTC=3600)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `SCHISMConfig` but got `SchismCSIROConfig` with value `SchismCSIROConfig(model_t...nput_history_DELTC=3600)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `VgridGenerator` with value `VgridGenerator(vgrid=Vgri..._type='LSC2_generator'))` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: PydanticSerializationUnexpectedValue: Expected `literal['vgrid2D_generator']` but got `str` with value `'LSC2_generator'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_LSC2` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_SZ` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `SchismCSIROMigrationConfig` but got `SchismCSIROConfig` with value `SchismCSIROConfig(model_t...nput_history_DELTC=3600)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `SwanConfig` but got `SchismCSIROConfig` with value `SchismCSIROConfig(model_t...nput_history_DELTC=3600)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `SwanConfigComponents` but got `SchismCSIROConfig` with value `SchismCSIROConfig(model_t...nput_history_DELTC=3600)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `SwanConfigComponents` but got `SchismCSIROConfig` with value `SchismCSIROConfig(model_t...nput_history_DELTC=3600)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `VgridGenerator` with value `VgridGenerator(vgrid=Vgri..._type='LSC2_generator'))` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: PydanticSerializationUnexpectedValue: Expected `literal['vgrid2D_generator']` but got `str` with value `'LSC2_generator'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_LSC2` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_SZ` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['vgrid2D_generator']` but got `str` with value `'LSC2_generator'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_LSC2` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_SZ` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `SCHISMDataBoundary` with value `SCHISMDataBoundary(model_...te_missing_coastal=True)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['boundary']` but got `str` with value `'data_boundary'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `SCHISMDataWave` with value `SCHISMDataWave(model_type...tion', data_type='wave')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['boundary']` but got `str` with value `'data_boundary'` - serialized value may not be as expected
Expected `dict[any, any]` but got `str` with value `'nearest'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `SCHISMDataTides` with value `SCHISMDataTides(data_type...[1], sobc=[1], relax=[])` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['tides']` but got `str` with value `'tide'` - serialized value may not be as expected
Expected `str` but got `int` with value `15` - serialized value may not be as expected
return self.__pydantic_serializer__.to_python(
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/pydantic/main.py:426: UserWarning: Pydantic serializer warnings:
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `VgridGenerator` with value `VgridGenerator(vgrid=Vgri..._type='LSC2_generator'))` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: PydanticSerializationUnexpectedValue: Expected `literal['vgrid2D_generator']` but got `str` with value `'LSC2_generator'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_LSC2` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_SZ` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['vgrid2D_generator']` but got `str` with value `'LSC2_generator'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_LSC2` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `Vgrid3D_SZ` but got `Vgrid2D` with value `Vgrid2D(model_type='LSC2_generator')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `SCHISMDataBoundary` with value `SCHISMDataBoundary(model_...te_missing_coastal=True)` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['boundary']` but got `str` with value `'data_boundary'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `SCHISMDataWave` with value `SCHISMDataWave(model_type...tion', data_type='wave')` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['boundary']` but got `str` with value `'data_boundary'` - serialized value may not be as expected
Expected `dict[any, any]` but got `str` with value `'nearest'` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `DataBlob` but got `SCHISMDataTides` with value `SCHISMDataTides(data_type...[1], sobc=[1], relax=[])` - serialized value may not be as expected
PydanticSerializationUnexpectedValue: Expected `literal['tides']` but got `str` with value `'tide'` - serialized value may not be as expected
Expected `str` but got `int` with value `15` - serialized value may not be as expected
return self.__pydantic_serializer__.to_python(
INFO:rompy.schism.grid:Generated albedo with constant value of 0.15
INFO:rompy.schism.grid:Generated diffmin with constant value of 1e-06
INFO:rompy.schism.grid:Generated diffmax with constant value of 1.0
INFO:rompy.schism.grid:Generated watertype with constant value of 1.0
INFO:rompy.schism.grid:Generated windrot_geo2proj with constant value of 0.0
INFO:rompy.schism.grid:Generated manning with constant value of 1.0
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_declaritive/test_schismcsiro/hgrid.ll
INFO:rompy.schism.grid:Linking hgrid.gr3 to schism_declaritive/test_schismcsiro/hgrid_WWM.gr3
INFO:rompy.schism.data:Fetching air_1
/home/tdurrant/source/rompy/rompy-base/rompy/core/data.py:304: UserWarning: Times can't be serialized faithfully to int64 with requested units 'days since 2023-01-01'. Serializing with units 'hours since 2023-01-01' instead. Set encoding['dtype'] to floating point dtype to serialize with units 'days since 2023-01-01'. Set encoding['units'] to 'hours since 2023-01-01' to silence this warning .
self.ds.to_netcdf(outfile)
INFO:rompy.schism.data:Fetching elev2D
INFO:rompy.schism.data:Fetching wavedata
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/intake_xarray/netcdf.py:92: UserWarning: The specified chunks separate the stored chunks along dimension "time" starting at index 1. This could degrade performance. Instead, consider rechunking after loading.
self._ds = _open_dataset(url, chunks=self.chunks, **kwargs)
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/intake_xarray/netcdf.py:92: UserWarning: The specified chunks separate the stored chunks along dimension "time" starting at index 1. This could degrade performance. Instead, consider rechunking after loading.
self._ds = _open_dataset(url, chunks=self.chunks, **kwargs)
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/intake_xarray/base.py:21: FutureWarning: The return type of `Dataset.dims` will be changed to return a set of dimension names in future, in order to be more consistent with `DataArray.dims`. To access a mapping from dimension names to lengths, please use `Dataset.sizes`.
'dims': dict(self._ds.dims),
/home/tdurrant/.virtualenvs/rompy-base-xqus/lib/python3.12/site-packages/wavespectra/output/ww3.py:115: UserWarning: Times can't be serialized faithfully to int64 with requested units 'days since 1990-01-01'. Resolution of 'hours' needed. Serializing times to floating point instead. Set encoding['dtype'] to integer dtype to serialize to int64. Set encoding['dtype'] to floating point dtype to silence this warning.
other.to_netcdf(filename)
INFO:rompy.schism.data:Generating tides
INFO:pyschism.forcing.bctides.bctides:Processing boundary 1:
INFO:pyschism.forcing.bctides.bctides:Elevation type: 5
WARNING:pyschism.forcing.bctides.bctides:Combination of 3 and 4, time history of elevation is read in from elev2D.th.nc!
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent M2.
INFO:pyschism.forcing.bctides.tpxo:h_file is ../../tests/schism/test_data/tpxo9-neaus/h_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for elevation constituent N2.
INFO:pyschism.forcing.bctides.bctides:Velocity type: 3
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent M2.
INFO:pyschism.forcing.bctides.tpxo:u_file is ../../tests/schism/test_data/tpxo9-neaus/u_m2s2n2.nc
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent S2.
INFO:pyschism.forcing.bctides.tpxo:Querying TPXO for velocity constituent N2.
INFO:pyschism.forcing.bctides.bctides:Temperature type: 0
WARNING:pyschism.forcing.bctides.bctides:Temperature is not sepcified, not input needed!
INFO:pyschism.forcing.bctides.bctides:Salinity type: 0
INFO:pyschism.forcing.bctides.bctides:Salinity is not sepcified, not input needed!
INFO:rompy.model:
INFO:rompy.model:Successfully generated project in schism_declaritive
INFO:rompy.model:-----------------------------------------------------