Basic demonstration#
This notebook it intended to demonstrate some of the basic concepts in the new version of of the rompy code
Separation of ‘runtime’ from ‘config’ - more on this later
Modular
There are a number of core objects
Pydantic throughout
Strong typing
Clearly defined APIs
Built in validators
Declaritive
The entire configuration to run the model is passed at intantiation.
The core wrapper is a pycallable, making it play nicely with downstream workflow packages (e.g. airflow)
Can still be run proceduraly.
Separation of intake components.
Common filters now available as a separate import.
Data is provdided to the wrapper as large lazy datasets, and data selection is done based on runtime period and grid spatial extents
Substantially reduced the fricton to hook up a different dataset
Filters are still available to use in intake drivers where appropriate (e.g. where constructing nowcasts from many forecasts becomes memory prohibitive to do lazily)
[1]:
# Set up notebook
%load_ext autoreload
%autoreload 2
%matplotlib inline
[2]:
# Import some core objects
from rompy import ModelRun
from rompy.core import TimeRange, BaseConfig, BaseGrid, RegularGrid, DatasetXarray
from rompy.swan import SwanConfig, SwanDataGrid, SwanGrid
from rompy.swan.data import SwanDataGrid
import xarray as xr
import numpy as np
import tempfile
import os
import pandas as pd
from datetime import datetime
import logging
logging.basicConfig(level=logging.INFO)
logger = logging.getLogger(__name__)
[3]:
# The highest level object is the ModelRun which is a generic class to run any model type.
# Lets start by looking at the ModelRun api
ModelRun?
Init signature:
ModelRun(
*,
run_id: str = 'run_id',
period: rompy.core.time.TimeRange = TimeRange(start=datetime.datetime(2020, 2, 21, 4, 0), end=datetime.datetime(2020, 2, 24, 4, 0), duration=datetime.timedelta(days=3), interval=datetime.timedelta(seconds=900), include_end=True),
output_dir: str = './simulations',
config: rompy.core.config.BaseConfig | rompy.swan.config.SwanConfig = BaseConfig(model_type='base', template='/source/rompy/rompy/templates/base', checkout='main'),
) -> None
Docstring:
A model run.
It is intented to be model agnostic.
It deals primarily with how the model is to be run, i.e. the period of the run
and where the output is going. The actual configuration of the run is
provided by the config object.
Further explanation is given in the rompy.core.Baseconfig docstring.
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/model.py
Type: ModelMetaclass
Subclasses:
[4]:
# So we can see that the ModelRun takes an id, a time run period, an output directort and a model configuration.
# TimeRange is a simple class that calculates run dates based on combinations of start, end, and interval.
TimeRange?
Init signature:
TimeRange(
*,
start: Optional[datetime.datetime] = None,
end: Optional[datetime.datetime] = None,
duration: Union[str, datetime.timedelta, NoneType] = None,
interval: Union[str, datetime.timedelta, NoneType] = '1h',
include_end: bool = True,
) -> None
Docstring:
A time range object
Examples
--------
>>> from rompy import TimeRange
>>> tr = TimeRange(start="2020-01-01", end="2020-01-02")
>>> tr
TimeRange(start=datetime.datetime(2020, 1, 1, 0, 0), end=datetime.datetime(2020, 1, 2, 0, 0), duration=None, interval=None, include_end=True)
>>> tr = TimeRange(start="2020-01-01", duration="1d")
>>> tr
TimeRange(start=datetime.datetime(2020, 1, 1, 0, 0), end=datetime.datetime(2020, 1, 2, 0, 0), duration=timedelta(days=1), interval=None, include_end=True)
>>> tr = TimeRange(start="2020-01-01", duration="1d", interval="1h")
>>> tr
TimeRange(start=datetime.datetime(2020, 1, 1, 0, 0), end=None, duration=timedelta(days=1), interval=timedelta(hours=1), include_end=True)
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/core/time.py
Type: ModelMetaclass
Subclasses:
[5]:
# The model configuration is a generic object that can be used to configure any model type.
# The simplest model configuration is the BaseConfig on which all model configurations are based.
BaseConfig??
Init signature:
BaseConfig(
*,
model_type: Literal['base'] = 'base',
template: Optional[str] = '/source/rompy/rompy/templates/base',
checkout: Optional[str] = 'main',
**extra_data: Any,
) -> None
Source:
class BaseConfig(RompyBaseModel):
"""Base class for model templates.
The template class provides the object that is used to set up the model configuration.
When implemented for a given model, can move along a scale of complexity
to suit the application.
In its most basic form, as implemented in this base object, it consists of path to a cookiecutter template
with the class providing the context for the {{config}} values in that template. Note that any
{{runtime}} values are filled from the ModelRun object.
If the template is a git repo, the checkout parameter can be used to specify a branch or tag and it
will be cloned and used.
If the object is callable, it will be colled prior to rendering the template. This mechanism can be
used to perform tasks such as fetching exteral data, or providing additional context to the template
beyond the arguments provided by the user..
"""
model_type: Literal["base"] = "base"
template: Optional[str] = Field(
description="The path to the model template",
default=DEFAULT_TEMPLATE,
)
checkout: Optional[str] = Field(
description="The git branch to use if the template is a git repo",
default="main",
)
class Config:
extra = "allow"
File: /source/rompy/rompy/core/config.py
Type: ModelMetaclass
Subclasses: SwanConfig, SwanConfigComponents
[6]:
# So a configuraton is just a dictionary of key value pairs coupled with a path to a template to used to render these arguments (more on this later).
# If the path to the template is a local file, this is used directly, if it points to a remote repository, that will be cloned locally using the
# cookiecutter mechanics and the path to the local copy will be used.
[7]:
# Before we start to look at the derived swan classes, lets look at a couple of other important base classes.
# First, a generic model grid
BaseGrid?
Init signature:
BaseGrid(
*,
x: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
y: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
grid_type: Literal['base'] = 'base',
) -> None
Docstring:
An object which provides an abstract representation of a grid in some geographic space
This is the base class for all Grid objects. The minimum representation of a grid are two
NumPy array's representing the vertices or nodes of some structured or unstructured grid,
its bounding box and a boundary polygon. No knowledge of the grid connectivity is expected.
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/core/grid.py
Type: ModelMetaclass
Subclasses: RegularGrid
[8]:
# Regular grids inherit from this base grid class, but can be defined by some simple paramaters due to their regular nature.
RegularGrid?
Init signature:
RegularGrid(
*,
x: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
y: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
grid_type: Literal['regular'] = 'regular',
x0: Optional[float] = None,
y0: Optional[float] = None,
rot: Optional[float] = 0.0,
dx: Optional[float] = None,
dy: Optional[float] = None,
nx: Optional[int] = None,
ny: Optional[int] = None,
) -> None
Docstring:
An object which provides an abstract representation of a regular grid in
some geographic space
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/core/grid.py
Type: ModelMetaclass
Subclasses: SwanGrid
[9]:
# Now lets look at some on the the swan specific classes that inherit from these base classes
from rompy.swan import SwanDataGrid, SwanGrid, SwanConfig
[10]:
# Swan grid inherits from regular grid (for now) and adds some swan specific parameters such as
# exc and implements swan grid inpgrid and cgrid methods
SwanGrid?
Init signature:
SwanGrid(
*,
x: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
y: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
grid_type: Literal['REG'] = 'REG',
x0: Optional[float] = None,
y0: Optional[float] = None,
rot: Optional[float] = 0.0,
dx: Optional[float] = None,
dy: Optional[float] = None,
nx: Optional[int] = None,
ny: Optional[int] = None,
exc: Optional[float] = None,
gridfile: Optional[rompy.swan.grid.ConstrainedStrValue] = None,
) -> None
Docstring:
An object which provides an abstract representation of a regular SWAN
grid in some geographic space
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/grid.py
Type: ModelMetaclass
Subclasses:
[11]:
SwanConfig?
Init signature:
SwanConfig(
*,
model_type: Literal['swan'] = 'swan',
template: str = '/source/rompy/rompy/templates/swan',
checkout: Optional[str] = 'main',
grid: rompy.swan.grid.SwanGrid,
spectral_resolution: rompy.swan.config.SwanSpectrum = SwanSpectrum(fmin=0.0464, fmax=1.0, nfreqs=31, ndirs=36),
forcing: rompy.swan.config.ForcingData = ForcingData(bottom=None, wind=None, current=None, boundary=None),
physics: rompy.swan.config.SwanPhysics = SwanPhysics(friction='MAD', friction_coeff=0.1),
outputs: rompy.swan.config.Outputs = Outputs(grid=GridOutput(period=None, variables=['DEPTH', 'UBOT', 'HSIGN', 'HSWELL', 'DIR', 'TPS', 'TM01', 'WIND']), spec=SpecOutput(period=None, locations=OutputLocs
)),
spectra_file: str = 'boundary.spec',
**extra_data: Any,
) -> None
Docstring: SWAN configuration
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/config.py
Type: ModelMetaclass
Subclasses:
[12]:
# So we can see that the SwanConfig is a BaseConfig with some additional arguments.
# These arguments are broken up into several compoents, lets examine these individually.
from rompy.swan.config import SwanSpectrum, ForcingData, SwanPhysics, Outputs, SwanDataGrid
[13]:
SwanSpectrum?
Init signature:
SwanSpectrum(
*,
fmin: float = 0.0464,
fmax: float = 1.0,
nfreqs: int = 31,
ndirs: int = 36,
) -> None
Docstring: SWAN Spectrum
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/config.py
Type: ModelMetaclass
Subclasses:
[14]:
SwanDataGrid?
Init signature:
SwanDataGrid(
*,
id: str,
dataset: rompy.core.data.DatasetXarray | rompy.core.data.DatasetIntake,
filter: Optional[rompy.core.filters.Filter] = Filter(sort={}, subset={}, crop={}, timenorm={}, rename={}, derived={}),
variables: Optional[list[str]] = [],
latname: Optional[str] = 'latitude',
lonname: Optional[str] = 'longitude',
timename: Optional[str] = 'time',
z1: str = None,
z2: str = None,
var: str = 'WIND',
) -> None
Docstring: This class is used to write SWAN data from a dataset.
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/data.py
Type: ModelMetaclass
Subclasses:
[15]:
SwanPhysics?
Init signature: SwanPhysics(*, friction: str = 'MAD', friction_coeff: float = 0.1) -> None
Docstring: Container class represting configuraable SWAN physics options
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/config.py
Type: ModelMetaclass
Subclasses:
[16]:
ForcingData?
Init signature:
ForcingData(
*,
bottom: rompy.swan.data.SwanDataGrid | None = None,
wind: rompy.swan.data.SwanDataGrid | None = None,
current: rompy.swan.data.SwanDataGrid | None = None,
boundary: rompy.swan.boundary.DataBoundary | None = None,
) -> None
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/config.py
Type: ModelMetaclass
Subclasses:
[17]:
SwanGrid?
Init signature:
SwanGrid(
*,
x: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
y: Optional[pydantic_numpy.ndarray.PydanticNDArray] = None,
grid_type: Literal['REG'] = 'REG',
x0: Optional[float] = None,
y0: Optional[float] = None,
rot: Optional[float] = 0.0,
dx: Optional[float] = None,
dy: Optional[float] = None,
nx: Optional[int] = None,
ny: Optional[int] = None,
exc: Optional[float] = None,
gridfile: Optional[rompy.swan.grid.ConstrainedStrValue] = None,
) -> None
Docstring:
An object which provides an abstract representation of a regular SWAN
grid in some geographic space
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/grid.py
Type: ModelMetaclass
Subclasses:
[18]:
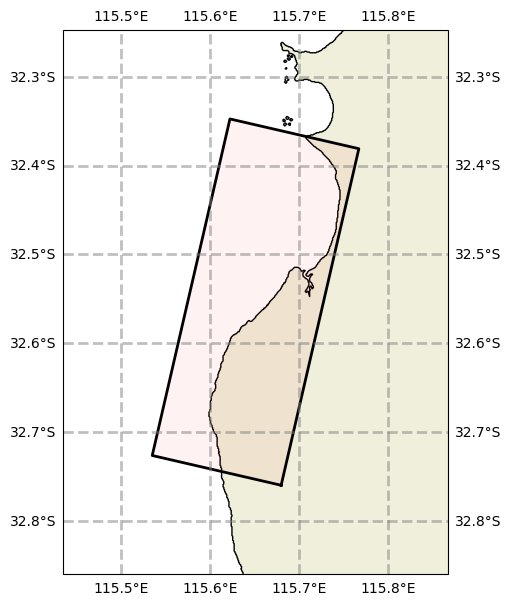
# Lets start by intantiaing a Swan grid
grid = SwanGrid(x0=115.68, y0=-32.76, dx=0.001, dy=0.001, nx=390, ny=150, rot=77)
grid.plot()
[18]:
(<Figure size 1000x706.373 with 1 Axes>, <GeoAxes: >)
[19]:
# Now lets look at how we might use that grid to define some data inputs
SwanDataGrid?
Init signature:
SwanDataGrid(
*,
id: str,
dataset: rompy.core.data.DatasetXarray | rompy.core.data.DatasetIntake,
filter: Optional[rompy.core.filters.Filter] = Filter(sort={}, subset={}, crop={}, timenorm={}, rename={}, derived={}),
variables: Optional[list[str]] = [],
latname: Optional[str] = 'latitude',
lonname: Optional[str] = 'longitude',
timename: Optional[str] = 'time',
z1: str = None,
z2: str = None,
var: str = 'WIND',
) -> None
Docstring: This class is used to write SWAN data from a dataset.
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/data.py
Type: ModelMetaclass
Subclasses:
[20]:
# Here we are creating some dummy data here to use as a bathymetry input
from rompy.swan import SwanDataGrid
os.makedirs("simulations/test_swantemplate/datasets", exist_ok=True)
datagrid = SwanGrid(x0=115.68, y0=-32.76, rot=77, nx=391, ny=151, dx=0.01, dy=0.01, exc=-99.0)
def nc_bathy(bottom=datagrid):
# touch temp netcdf file
tmp_path = tempfile.mkdtemp()
source = os.path.join("simulations", "test_swantemplate", "datasets", "bathy.nc")
bbox = bottom.bbox(buffer=1)
lat = np.arange(bbox[1], bbox[3], bottom.dy)
lon = np.arange(bbox[0], bbox[2], bottom.dx)
ds = xr.Dataset(
{
"depth": xr.DataArray(
np.random.rand(lat.size, lon.size),
dims=["lat", "lon"],
coords={"lat": lat, "lon": lon},
),
}
)
ds.to_netcdf(source, mode="w")
return source
# and same again for wind
def nc_data_source(wind_grid=datagrid):
tmp_path = tempfile.mkdtemp()
source = os.path.join("simulations", "test_swantemplate", "datasets", "wind_inputs.nc")
bbox = wind_grid.bbox(buffer=1)
lat = np.arange(bbox[1], bbox[3], wind_grid.dy)
lon = np.arange(bbox[0], bbox[2], wind_grid.dx)
time = pd.date_range("2023-01-01", periods=10)
ds = xr.Dataset(
{
"u": xr.DataArray(
np.random.rand(10, lat.size, lon.size),
dims=["time", "lat", "lon"],
coords={
"time": time,
"lat": lat,
"lon": lon,
},
),
"v": xr.DataArray(
np.random.rand(10, lat.size, lon.size),
dims=["time", "lat", "lon"],
coords={
"time": time,
"lat": lat,
"lon": lon,
},
),
}
)
ds.to_netcdf(source, mode="w")
return source
[21]:
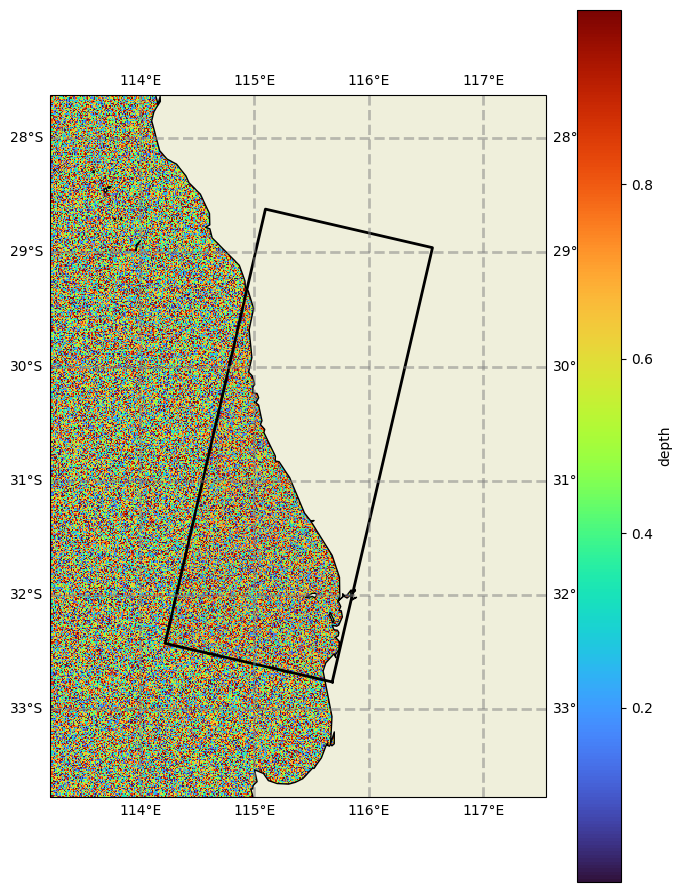
# Create dummy data
bathy_source = nc_bathy()
# Intantiate a swan data object using this dummy data as input
bathy = SwanDataGrid(
id="bottom", dataset=DatasetXarray(uri=bathy_source), z1="depth", var="BOTTOM", latname="lat", lonname="lon"
)
# Plot the full dataset
bathy.plot(param='depth', fscale=8, model_grid=datagrid)
[21]:
(<Figure size 800x1132.56 with 2 Axes>, <GeoAxes: xlabel='lon', ylabel='lat'>)
[22]:
# If we run the bathy get method, this will write the data in swan format to the stage directory ready for ingestion into the model.
# At the moment, this is the full dataset, but we will look at subsetting options later
# Note that running this method retuns the required control file commands to read the data
# from the control file
bathy.get('./')
INFO:rompy.swan.data: Writing bottom to ./bottom.grd
[22]:
"INPGRID BOTTOM REG 113.21844490282216 -33.76 0.0 433 613 0.010000000000005116 0.00999999999999801 EXC -99.0\nREADINP BOTTOM 1.0 'bottom.grd' 3 FREE\n"
[23]:
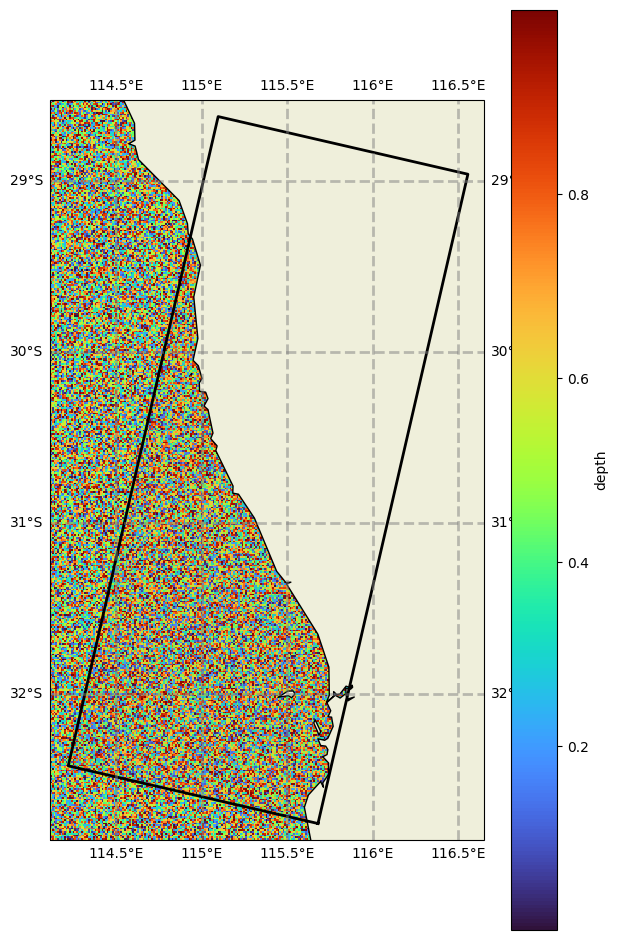
# But instead of just using the data as is, we can use filters to subset it. There are a number of filters available.
# Here we just demonstrate some hidden convenience functions which use a model grid to subset space.
bathy._filter_grid(datagrid)
bathy.get('./')
bathy.plot(param='depth', fscale=7, model_grid=datagrid)
INFO:rompy.swan.data: Writing bottom to ./bottom.grd
[23]:
(<Figure size 700x1195.26 with 2 Axes>, <GeoAxes: xlabel='lon', ylabel='lat'>)
[24]:
# This has now extracted only what it needs based on the grid extents.
# This is what is done internally in the wrapper (in addition to a analogous time filtering function) allowing us to use lazy grids without doing any pre-filtering,
# and have the wrapper use only what it needs
[25]:
# Wind data works the same way. Note the control file input commands are different as it now applied to wind data
wind_source = nc_data_source()
wind = SwanDataGrid(id="wind", var="WIND", dataset=DatasetXarray(uri=wind_source), z1="u", z2="v")
wind.get('./')
INFO:rompy.swan.data: Writing wind to ./wind.grd
[25]:
"INPGRID WIND REG 113.21844490282216 -33.76 0.0 433 613 0.010000000000005116 0.00999999999999801 NONSTATION 20230101.000000 24.0 HR\nREADINP WIND 1 'wind.grd' 3 0 1 0 FREE\n"
[26]:
# The boundary object work in a similar way but is a little more complex as it must determine points around the boundary of the model domain.
# First lets have a look at the boundary object
from rompy.swan import Boundnest1
Boundnest1?
Init signature:
DataBoundary(
*,
id: str,
dataset: rompy.core.data.DatasetXarray | rompy.core.data.DatasetIntake | rompy.swan.boundary.DatasetWavespectra,
filter: Optional[rompy.core.filters.Filter] = Filter(sort={}, subset={}, crop={}, timenorm={}, rename={}, derived={}),
variables: Optional[list[str]] = [],
latname: Optional[str] = 'latitude',
lonname: Optional[str] = 'longitude',
timename: Optional[str] = 'time',
spacing: Optional[float] = None,
sel_method: Literal['idw', 'nearest'] = 'idw',
tolerance: rompy.swan.boundary.ConstrainedFloatValue = 1.0,
rectangle: Literal['closed', 'open'] = 'closed',
) -> None
Docstring:
SWAN BOUNDNEST1 NEST data class.
Notes
-----
The `tolerance` behaves differently with sel_methods `idw` and `nearest`; in `idw`
sites with no enough neighbours within `tolerance` are masked whereas in `nearest`
an exception is raised (see wavespectra documentation for more details).
Be aware that when using `idw` missing values will be returned for sites with less
than 2 neighbours within `tolerance` in the original dataset. This is okay for land
mask areas but could cause boundary issues when on an open boundary location. To
avoid this either use `nearest` or increase `tolerance` to include more neighbours.
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/boundary.py
Type: ModelMetaclass
Subclasses:
[27]:
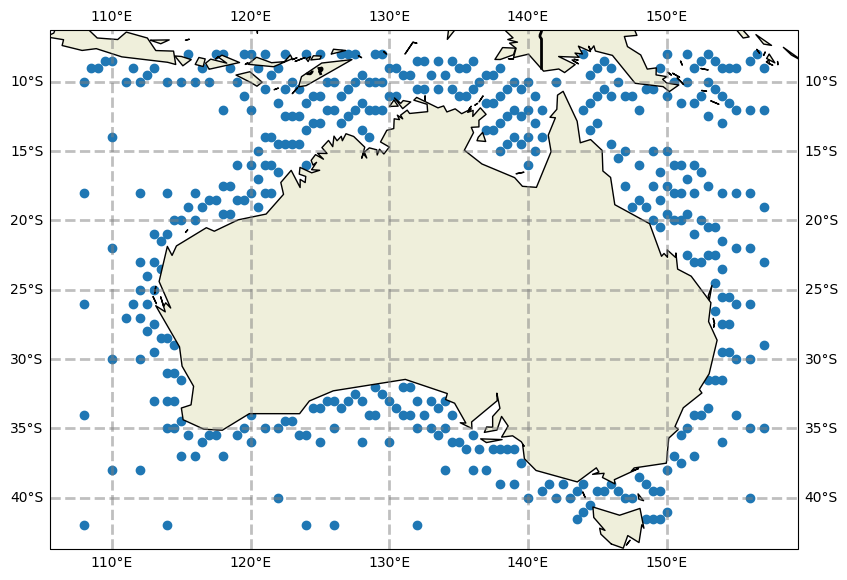
# Here we use a test boundary dataset
# Instantiate the boundary object
bnd = DataBoundary(id='bnd', dataset=DatasetXarray(uri="../tests/data/aus-20230101.nc"), latname='lat', lonname='lon', time='time', tolerance=0.1, sel_method='idw')
# plot the unfiltered data locations
bnd.plot(model_grid=grid)
#bnd.ds
[28]:
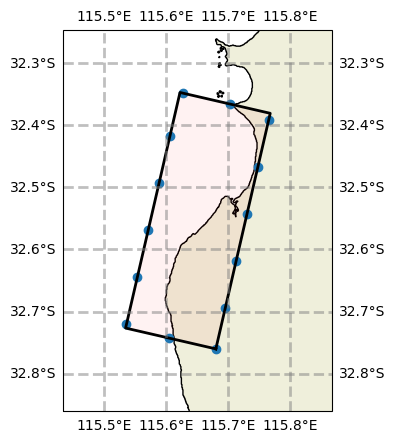
# Now we can filter the boundary data to the model grid. Here we just use a convenience function to plot the results of this effort
# TODO - a bit more exlanation needed here
bnd.plot_boundary(grid, fscale=7)
[29]:
# Putting these together we can create a swan forcing object When used in a model run, this object will loop over all the data source, filter in time and space to match the model run,
# write the data to the scratch space, and produce the required control file inputs for swan
forcing = ForcingData(wind=wind, bottom=bathy, boundary=bnd)
forcing.get?
Signature:
forcing.get(
grid: rompy.swan.grid.SwanGrid,
period: rompy.core.time.TimeRange,
staging_dir: pathlib.Path,
)
Docstring: <no docstring>
File: /source/rompy/rompy/swan/config.py
Type: method
[30]:
# Next, lets look at the physics object. At the moment, this is very simple and only
# templates friction in line with the old rompy, but this is now very easy to
# extend to other physics options
SwanPhysics?
Init signature: SwanPhysics(*, friction: str = 'MAD', friction_coeff: float = 0.1) -> None
Docstring: Container class represting configuraable SWAN physics options
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/config.py
Type: ModelMetaclass
Subclasses:
[31]:
# Just as a demonstration of a custom validation error, lets try to instantiate a physics object with an invalid friction option
try:
physics = SwanPhysics(friction='INVALID')
except ValueError as e:
logger.warning(e)
WARNING:__main__:1 validation error for SwanPhysics
friction
friction must be one of JON, COLL, MAD or RIP (type=value_error)
[32]:
# Now lets instantiate a valid physics object
physics = SwanPhysics()
# And render its cmd file method
print(physics.cmd)
GEN3 WESTH 0.000075 0.00175
BREAKING
FRICTION MAD 0.1
TRIADS
PROP BSBT
NUM ACCUR 0.02 0.02 0.02 95 NONSTAT 20
[33]:
# Finally, the outputs object is used to define the outputs of the model run.
# These specify grid variables and spectral locations to be outputted.
# They also define a period for output, which defaults to the full model run
Outputs?
Init signature:
Outputs(
*,
grid: rompy.swan.config.GridOutput = GridOutput(period=None, variables=['DEPTH', 'UBOT', 'HSIGN', 'HSWELL', 'DIR', 'TPS', 'TM01', 'WIND']),
spec: rompy.swan.config.SpecOutput = SpecOutput(period=None, locations=OutputLocs
),
) -> None
Docstring: Outputs for SWAN
Init docstring:
Create a new model by parsing and validating input data from keyword arguments.
Raises ValidationError if the input data cannot be parsed to form a valid model.
File: /source/rompy/rompy/swan/config.py
Type: ModelMetaclass
Subclasses:
[34]:
# TODO fix below
#outputs = Outputs(spec=SpecOutput(locations=[[115.68, -32.76]]), period=TimeRange(start=datetime(2020, 2, 21), end=datetime(2020, 2, 22)))
# Instantiate default model output
outputs = Outputs()
outputs
[34]:
Outputs(grid=GridOutput(period=None, variables=['DEPTH', 'UBOT', 'HSIGN', 'HSWELL', 'DIR', 'TPS', 'TM01', 'WIND']), spec=SpecOutput(period=None, locations=OutputLocs
))
[35]:
# Lets now put those components into a swan config object and instantiate a model run
run = ModelRun(
run_id="test_swantemplate",
period=TimeRange(start=datetime(2023, 1, 1, 0), end=datetime(2023, 1, 4, 4)),
output_dir="simulations",
config=SwanConfig(
grid=grid,
physics=physics,
forcing=forcing,
outputs=outputs,
)
)
[36]:
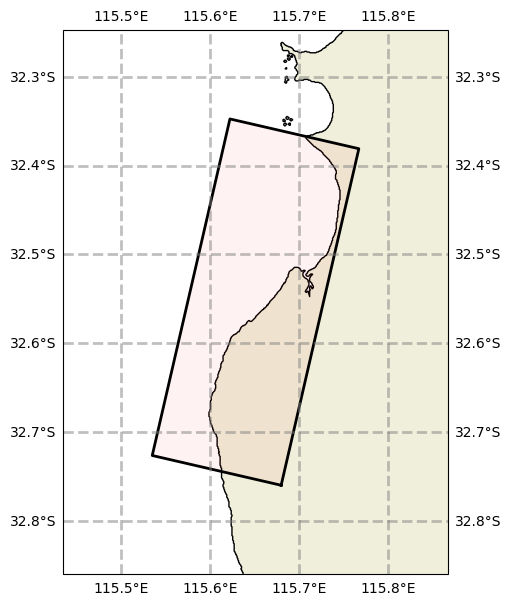
# This grid object is still accessible as before, but the path has changed slightly
run.config.grid.plot()
[36]:
(<Figure size 1000x706.373 with 1 Axes>, <GeoAxes: >)
[37]:
# And we can also run individual components and look at the control file inputs
cmd = run.config.forcing.get(grid=run.config.grid, runtime=run)
print(cmd['forcing'])
print(cmd['boundary'])
---------------------------------------------------------------------------
TypeError Traceback (most recent call last)
Cell In[37], line 2
1 # And we can also run individual components and look at the control file inputs
----> 2 cmd = run.config.forcing.get(grid=run.config.grid, runtime=run)
3 print(cmd['forcing'])
4 print(cmd['boundary'])
TypeError: ForcingData.get() got an unexpected keyword argument 'runtime'
[ ]:
run.config.outputs.spec.locations
OutputLocs
[ ]:
run()
INFO:rompy.model:
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Model settings:
INFO:rompy.model:
period:
Start: 2023-01-01 00:00:00
End: 2023-01-04 04:00:00
Duration: 3 days, 4:00:00
Interval: 1:00:00
Include End: True
output_dir:
simulations
config:
grid:
SwanGrid: REG, 390x150
spectral_resolution:
fmin=0.0464 fmax=1.0 nfreqs=31 ndirs=36
forcing:
bottom: DatasetXarray(uri=simulations/test_swantemplate/datasets/bathy.nc
wind: DatasetXarray(uri=simulations/test_swantemplate/datasets/wind_inputs.nc
boundary: DatasetXarray(uri=../tests/data/aus-20230101.nc
physics:
friction='MAD' friction_coeff=0.1
outputs:
Grid:
variables: DEPTH UBOT HSIGN HSWELL DIR TPS TM01 WIND
Spec
locations:
template:
/source/rompy/rompy/templates/swan
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Generating model input files in simulations
INFO:rompy.swan.config: Processing bottom forcing
INFO:rompy.swan.data: Writing bottom to simulations/test_swantemplate/bottom.grd
INFO:rompy.swan.config: Processing wind forcing
INFO:rompy.swan.data: Writing wind to simulations/test_swantemplate/wind.grd
INFO:rompy.swan.config: Processing boundary forcing
INFO:rompy.model:
INFO:rompy.model:Successfully generated project in simulations
INFO:rompy.model:-----------------------------------------------------
'/source/rompy/notebooks/simulations/test_swantemplate'
Command line interface#
[ ]:
# Another advantage of using a declaritive model is that configuration becomes much easier to manage.
# We have steped through each object separately, but the whole model run can be described with simple
# arguments. To illustrate this, lets load a simple configuration file to describes the full model run above.
!cat demo.yml
# Note that this is config fully replicates the config deomonstrated in the demo notebook,
# however it requireds the datasets created in that notebook to work.
run_id: test_swantemplate
period:
start: 20230101T00
# end: 20200224T04
duration: 3d
interval: 1h
output_dir: simulations
config:
model_type: swan
grid:
x0: 115.68
y0: -32.76
rot: 77.0
dx: 0.001
dy: 0.001
nx: 390
ny: 150
gridtype: REG
spectral_resolution:
fmin: 0.0464
fmax: 1.0
nfreqs: 31
ndirs: 36
forcing:
wind:
model_type: swan
id: wind
var: WIND
dataset:
uri: simulations/test_swantemplate/datasets/wind_inputs.nc
model_type: xarray
z1: u
z2: v
latname: lat
lonname: lon
bottom:
id: bottom
var: BOTTOM
dataset:
uri: simulations/test_swantemplate/datasets/bathy.nc
model_type: xarray
z1: depth
latname: lat
lonname: lon
boundary:
id: bnd
dataset:
uri: ../tests/data/aus-20230101.nc
model_type: xarray
latname: lat
lonname: lon
tolerance: 0.1
sel_method: idw
physics':
friction: MAD
friction_coeff: 0.1
outputs:
grid:
variables:
- DEPTH
- UBOT
- HSIGN
- HSWELL
- DIR
- TPS
- TM01
- WIND
spec:
locations:
- x: 115.68
y: -32.76
- x: 115.68
y: -32.76
[ ]:
# initantiating a model run using this config
import yaml
args = yaml.load(open('demo.yml', 'r'), Loader=yaml.FullLoader)
import wavespectra
run = ModelRun(**args)
# and then calling as before
run()
---------------------------------------------------------------------------
ValidationError Traceback (most recent call last)
Cell In[80], line 5
3 args = yaml.load(open('demo.yml', 'r'), Loader=yaml.FullLoader)
4 import wavespectra
----> 5 run = ModelRun(**args)
7 # and then calling as before
8 run()
File /source/rompy/venv/lib/python3.10/site-packages/pydantic/main.py:341, in pydantic.main.BaseModel.__init__()
ValidationError: 1 validation error for ModelRun
config -> SwanConfig
descriptor '_datefmt' for 'SwanConfig' objects doesn't apply to a 'SwanConfig' object (type=type_error)
[ ]:
# There is also a command line interface to run defined model configuration
!rompy --help
Usage: rompy [OPTIONS] {base|swan} CONFIG
Run model Usage: rompy <model> config.yml Args: model(str): model type
config(str): yaml config file
Options:
-k, --kwargs TEXT additional key value pairs in the format key:value
--help Show this message and exit.
[ ]:
!rompy swan demo.yml
INFO:rompy.model:
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Model settings:
INFO:rompy.model:
period:
Start: 2023-01-01 00:00:00
End: 2023-01-04 00:00:00
Duration: 3 days, 0:00:00
Interval: 1:00:00
Include End: True
output_dir:
simulations
config:
grid:
SwanGrid: REG, 390x150
spectral_resolution:
fmin=0.0464 fmax=1.0 nfreqs=31 ndirs=36
forcing:
bottom: DatasetXarray(uri=simulations/test_swantemplate/datasets/bathy.nc
wind: DatasetXarray(uri=simulations/test_swantemplate/datasets/wind_inputs.nc
boundary: DatasetXarray(uri=../tests/data/aus-20230101.nc
physics:
friction='MAD' friction_coeff=0.1
outputs:
Grid:
variables: DEPTH UBOT HSIGN HSWELL DIR TPS TM01 WIND
Spec
locations:
template:
/source/rompy/rompy/templates/swan
INFO:rompy.model:-----------------------------------------------------
INFO:rompy.model:Generating model input files in simulations
INFO:rompy.swan.config: Processing bottom forcing
INFO:rompy.swan.data: Writing bottom to simulations/test_swantemplate/bottom.grd
INFO:rompy.swan.config: Processing wind forcing
INFO:rompy.swan.data: Writing wind to simulations/test_swantemplate/wind.grd
INFO:rompy.swan.config: Processing boundary forcing
INFO:rompy.model:
INFO:rompy.model:Successfully generated project in simulations
INFO:rompy.model:-----------------------------------------------------
[ ]:
# So hopefully that gives a the basic functionality. A bit more detail on templates is given in template_demo.ipynb